Cloning of Beauveria bassiana chitinase gene Bbchit1 and its application to improve fungal strain virulence
- PMID: 15640210
- PMCID: PMC544255
- DOI: 10.1128/AEM.71.1.363-370.2005
Cloning of Beauveria bassiana chitinase gene Bbchit1 and its application to improve fungal strain virulence
Abstract
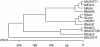
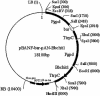
Entomopathogenic fungi can produce a series of chitinases, some of which act synergistically with proteases to degrade insect cuticle. However, chitinase involvement in insect fungus pathogenesis has not been fully characterized. In this paper, an endochitinase, Bbchit1, was purified to homogeneity from liquid cultures of Beauveria bassiana grown in a medium containing colloidal chitin. Bbchit1 had a molecular mass of about 33 kDa and pI of 5.4. Based on the N-terminal amino acid sequence, the chitinase gene, Bbchit1, and its upstream regulatory sequence were cloned. Bbchit1 was intronless, and there was a single copy in B. bassiana. Its regulatory sequence contained putative CreA/Crel carbon catabolic repressor binding domains, which was consistent with glucose suppression of Bbchit1. At the amino acid level, Bbchit1 showed significant similarity to a Streptomyces avermitilis putative endochitinase, a Streptomyces coelicolor putative chitinase, and Trichoderma harzianum endochitinase Chit36Y. However, Bbchit1 had very low levels of identity to other chitinase genes previously isolated from entomopathogenic fungi, indicating that Bbchit1 was a novel chitinase gene from an insect-pathogenic fungus. A gpd-Bbchit1 construct, in which Bbchit1 was driven by the Aspergiullus nidulans constitutive promoter, was transformed into the genome of B. bassiana, and three transformants that overproduced Bbchit1 were obtained. Insect bioassays revealed that overproduction of Bbchit1 enhanced the virulence of B. bassiana for aphids, as indicated by significantly lower 50% lethal concentrations and 50% lethal times of the transformants compared to the values for the wild-type strain.
Figures


References
-
- Bailey, C., and H. N. Arst, Jr. 1975. Carbon catabolite repression in Aspergillus nidulans. Eur. J. Biochem. 51:573-577. - PubMed
-
- Bidochka, M. J. 2001. Monitoring the fate of biocontrol fungi, p. 193-218. In T. M. Butt, C. Jackson, and N. Magan (ed.), Fungi as biocontrol agents. CABI, Wallingford, Oxon, United Kingdom.
-
- Bogo, M. R., C. A. Rota, H. Pinto, Jr., M. Ocampos, C. T. Correa, H. M. ainstein, and A. Schrank. 1998. A chitinase encoding gene (chit1 gene) from the entomopathogen Metarhizium anisopliae: isolation and characterization of genomic and full-length cDNA. Curr. Microbiol. 37:221-225. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

