Computational prediction of human metabolic pathways from the complete human genome
- PMID: 15642094
- PMCID: PMC549063
- DOI: 10.1186/gb-2004-6-1-r2
Computational prediction of human metabolic pathways from the complete human genome
Abstract
Background: We present a computational pathway analysis of the human genome that assigns enzymes encoded therein to predicted metabolic pathways. Pathway assignments place genes in their larger biological context, and are a necessary first step toward quantitative modeling of metabolism.
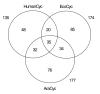
Results: Our analysis assigns 2,709 human enzymes to 896 bioreactions; 622 of the enzymes are assigned roles in 135 predicted metabolic pathways. The predicted pathways closely match the known nutritional requirements of humans. This analysis identifies probable omissions in the human genome annotation in the form of 203 pathway holes (missing enzymes within the predicted pathways). We have identified putative genes to fill 25 of these holes. The predicted human metabolic map is described by a Pathway/Genome Database called HumanCyc, which is available at http://HumanCyc.org/. We describe the generation of HumanCyc, and present an analysis of the human metabolic map. For example, we compare the predicted human metabolic pathway complement to the pathways of Escherichia coli and Arabidopsis thaliana and identify 35 pathways that are shared among all three organisms.
Conclusions: Our analysis elucidates a significant portion of the human metabolic map, and also indicates probable unidentified genes in the genome. HumanCyc provides a genome-based view of human nutrition that associates the essential dietary requirements of humans with a set of metabolic pathways whose existence is supported by the human genome. The database places many human genes in a pathway context, thereby facilitating analysis of gene expression, proteomics, and metabolomics datasets through a publicly available online tool called the Omics Viewer.
Figures


References
-
- Karp PD, Paley S, Romero P. The Pathway Tools software. Bioinformatics. 2002;18:S225–S232. - PubMed
-
- EcoCyc: encyclopedia of Escherichia coli K12 genes and metabolism. http://EcoCyc.org
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

