Wound healing and inflammation genes revealed by array analysis of 'macrophageless' PU.1 null mice
- PMID: 15642097
- PMCID: PMC549066
- DOI: 10.1186/gb-2004-6-1-r5
Wound healing and inflammation genes revealed by array analysis of 'macrophageless' PU.1 null mice
Abstract
Background: Wound healing is a complex process requiring the collaborative efforts of different tissues and cell lineages, and involving the coordinated interplay of several phases of proliferation, migration, matrix synthesis and contraction. Tissue damage also triggers a robust influx of inflammatory leukocytes to the wound site that play key roles in clearing the wound of invading microbes but also release signals that may be detrimental to repair and lead to fibrosis.
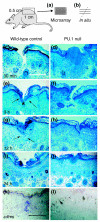
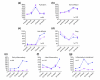
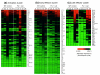
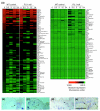
Results: To better define key cellular events pivotal for tissue repair yet independent of inflammation we have used a microarray approach to determine a portfolio of over 1,000 genes expressed across the repair response in a wild-type neonatal mouse versus its PU.1 null sib. The PU.1 null mouse is genetically incapable of raising the standard inflammatory response, because it lacks macrophages and functioning neutrophils, yet repairs skin wounds rapidly and with reduced fibrosis. Conversely, by subtraction, we have determined genes that are either expressed by leukocytes, or upregulated by fibroblasts, endothelial cells, muscle cells and others at the wound site, as a consequence of inflammation. To determine the spatial expression pattern for several genes in each cluster we have also performed in situ hybridization studies.
Conclusions: Cluster analysis of genes expressed after wounding wild-type mice versus PU.1 null sibs distinguishes between tissue repair genes and genes associated with inflammation and its consequences. Our data reveal and classify several pools of genes, giving insight into their likely functions during repair and hinting at potential therapeutic targets.
Figures


References
-
- Werner S, Grose R. Regulation of wound healing by growth factors and cytokines. Physiol Rev. 2003;83:835–870. - PubMed
-
- Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, Chi JT, Rijn Mv M, Botstein D, Brown PO. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020074. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

