Critical evaluation of the JDO API for the persistence and portability requirements of complex biological databases
- PMID: 15642112
- PMCID: PMC545948
- DOI: 10.1186/1471-2105-6-5
Critical evaluation of the JDO API for the persistence and portability requirements of complex biological databases
Abstract
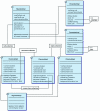
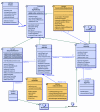
Background: Complex biological database systems have become key computational tools used daily by scientists and researchers. Many of these systems must be capable of executing on multiple different hardware and software configurations and are also often made available to users via the Internet. We have used the Java Data Object (JDO) persistence technology to develop the database layer of such a system known as the SigPath information management system. SigPath is an example of a complex biological database that needs to store various types of information connected by many relationships.
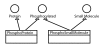
Results: Using this system as an example, we perform a critical evaluation of current JDO technology; discuss the suitability of the JDO standard to achieve portability, scalability and performance. We show that JDO supports portability of the SigPath system from a relational database backend to an object database backend and achieves acceptable scalability. To answer the performance question, we have created the SigPath JDO application benchmark that we distribute under the Gnu General Public License. This benchmark can be used as an example of using JDO technology to create a complex biological database and makes it possible for vendors and users of the technology to evaluate the performance of other JDO implementations for similar applications.
Conclusions: The SigPath JDO benchmark and our discussion of JDO technology in the context of biological databases will be useful to bioinformaticians who design new complex biological databases and aim to create systems that can be ported easily to a variety of database backends.
Figures



References
-
- Taylor CF, Paton NW, Garwood KL, Kirby PD, Stead DA, Yin Z, Deutsch EW, Selway L, Walker J, Riba-Garcia I, Mohammed S, Deery MJ, Howard JA, Dunkley T, Aebersold R, Kell DB, Lilley KS, Roepstorff P, Yates JR, Brass A, Brown AJ, Cash P, Gaskell SJ, Hubbard SJ, Oliver SG. A systematic approach to modeling, capturing, and disseminating proteomics experimental data. Nat Biotechnol. 2003;21:247–254. doi: 10.1038/nbt0303-247. - DOI - PubMed
-
- Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, Durbin R, Eyras E, Gilbert J, Hammond M, Huminiecki L, Kasprzyk A, Lehvaslaiho H, Lijnzaad P, Melsopp C, Mongin E, Pettett R, Pocock M, Potter S, Rust A, Schmidt E, Searle S, Slater G, Smith J, Spooner W, Stabenau A, Stalker J, Stupka E, Ureta-Vidal A, Vastrik I, Clamp M. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. - DOI - PMC - PubMed
-
- Oliver DE, Rubin DL, Stuart JM, Hewett M, Klein TE, Altman RB. Ontology development for a pharmacogenetics knowledge base. Pac Symp Biocomput. 2002:65–76. - PubMed
-
- Rubin DL, Hewett M, Oliver DE, Klein TE, Altman RB. Automating data acquisition into ontologies from pharmacogenetics relational data sources using declarative object definitions and XML. Pac Symp Biocomput. 2002:88–99. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

