Structure and evolution of the mouse pregnancy-specific glycoprotein (Psg) gene locus
- PMID: 15647114
- PMCID: PMC546212
- DOI: 10.1186/1471-2164-6-4
Structure and evolution of the mouse pregnancy-specific glycoprotein (Psg) gene locus
Abstract
Background: The pregnancy-specific glycoprotein (Psg) genes encode proteins of unknown function, and are members of the carcinoembryonic antigen (Cea) gene family, which is a member of the immunoglobulin gene (Ig) superfamily. In rodents and primates, but not in artiodactyls (even-toed ungulates / hoofed mammals), there have been independent expansions of the Psg gene family, with all members expressed exclusively in placental trophoblast cells. For the mouse Psg genes, we sought to determine the genomic organisation of the locus, the expression profiles of the various family members, and the evolution of exon structure, to attempt to reconstruct the evolutionary history of this locus, and to determine whether expansion of the gene family has been driven by selection for increased gene dosage, or diversification of function.
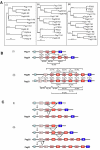
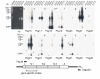
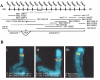
Results: We collated the mouse Psg gene sequences currently in the public genome and expressed-sequence tag (EST) databases and used systematic BLAST searches to generate complete sequences for all known mouse Psg genes. We identified a novel family member, Psg31, which is similar to Psg30 but, uniquely amongst mouse Psg genes, has a duplicated N1 domain. We also identified a novel splice variant of Psg16 (bCEA). We show that Psg24 and Psg30 / Psg31 have independently undergone expansion of N-domain number. By mapping BAC, YAC and cosmid clones we described two clusters of Psg genes, which we linked and oriented using fluorescent in situ hybridisation (FISH). Comparison of our Psg locus map with the public mouse genome database indicates good agreement in overall structure and further elucidates gene order. Expression levels of Psg genes in placentas of different developmental stages revealed dramatic differences in the developmental expression profile of individual family members.
Conclusion: We have combined existing information, and provide new information concerning the evolution of mouse Psg exon organization, the mouse Psg genomic locus structure, and the expression patterns of individual Psg genes. This information will facilitate functional studies of this complex gene family.
Figures



References
-
- Lei KJ, Sartwell AD, Pan CJ, Chou JY. Cloning and expression of genes encoding human pregnancy-specific glycoproteins. Journal of Biological Chemistry. 1992;267:16371–16378. - PubMed
-
- Rebstock S, Lucas K, Weiss M, Thompson J, Zimmermann W. Spatiotemporal expression of pregnancy-specific glycoprotein gene rnCGM1 in rat placenta. Developmental Dynamics. 1993;198:171–181. - PubMed
-
- Kromer B, Finkenzeller D, Wessels J, Dveksler G, Thompson J, Zimmermann W. Coordinate expression of splice variants of the murine pregnancy-specific glycoprotein (PSG) gene family during placental development. European Journal of Biochemistry. 1996;242:280–287. doi: 10.1111/j.1432-1033.1996.0280r.x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

