Genome-wide and Ordered-Subset linkage analyses provide support for autism loci on 17q and 19p with evidence of phenotypic and interlocus genetic correlates
- PMID: 15647115
- PMCID: PMC546213
- DOI: 10.1186/1471-2350-6-1
Genome-wide and Ordered-Subset linkage analyses provide support for autism loci on 17q and 19p with evidence of phenotypic and interlocus genetic correlates
Abstract
Background: Autism is a neurobehavioral spectrum of phenotypes characterized by deficits in the development of language and social relationships and patterns of repetitive, rigid and compulsive behaviors. Twin and family studies point to a significant genetic etiology, and several groups have performed genomic linkage screens to identify susceptibility loci.
Methods: We performed a genome-wide linkage screen in 158 combined Tufts, Vanderbilt and AGRE (Autism Genetics Research Exchange) multiplex autism families using parametric and nonparametric methods with a categorical autism diagnosis to identify loci of main effect. Hypothesizing interdependence of genetic risk factors prompted us to perform exploratory studies applying the Ordered-Subset Analysis (OSA) approach using LOD scores as the trait covariate for ranking families. We employed OSA to test for interlocus correlations between loci with LOD scores > or =1.5, and empirically determined significance of linkage in optimal OSA subsets using permutation testing. Exploring phenotypic correlates as the basis for linkage increases involved comparison of mean scores for quantitative trait-based subsets of autism between optimal subsets and the remaining families.
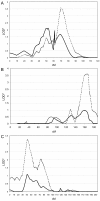
Results: A genome-wide screen for autism loci identified the best evidence for linkage to 17q11.2 and 19p13, with maximum multipoint heterogeneity LOD scores of 2.9 and 2.6, respectively. Suggestive linkage (LOD scores > or =1.5) at other loci included 3p, 6q, 7q, 12p, and 16p. OSA revealed positive correlations of linkage between the 19p locus and 17q, between 19p and 6q, and between 7q and 5p. While potential phenotypic correlates for these findings were not identified for the chromosome 7/5 combination, differences indicating more rapid achievement of "developmental milestones" was apparent in the chromosome 19 OSA-defined subsets for 17q and 6q. OSA was used to test the hypothesis that 19p linkage involved more rapid achievement of these milestones and it revealed significantly increased LOD* scores at 19p13.
Conclusions: Our results further support 19p13 as harboring an autism susceptibility locus, confirm other linkage findings at 17q11.2, and demonstrate the need to analyze more discreet trait-based subsets of complex phenotypes to improve ability to detect genetic effects.
Figures


References
-
- Volkmar FR, Szatmari P, Sparrow SS. Sex differences in pervasive developmental disorders. J Autism Dev Disord. 1993;23:579–591. - PubMed
-
- McLennan JD, Lord C, Schopler E. Sex differences in higher functioning people with autism. J Autism Dev Disord. 1993;23:217–227. - PubMed
-
- Folstein S, Rutter M. Infantile autism: a genetic study of 21 twin pairs. J Child Psychol Psychiatry. 1977;18:297–321. - PubMed
-
- Rutter M, Macdonald H, Le Couteur A, Harrington R, Bolton P, Bailey A. Genetic factors in child psychiatric disorders – ll. Empirical findings. J Child Psychol Psychiatry. 1990;31:39–83. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

