Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery
- PMID: 15647345
- PMCID: PMC545576
- DOI: 10.1073/pnas.0408782102
Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery
Abstract
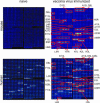
Despite the increasing availability of genome sequences from many human pathogens, the production of complete proteomes remains at a bottleneck. To address this need, a high-throughput PCR recombination cloning and expression platform has been developed that allows hundreds of genes to be batch-processed by using ordinary laboratory procedures without robotics. The method relies on high-throughput amplification of each predicted ORF by using gene specific primers, followed by in vivo homologous recombination into a T7 expression vector. The proteins are expressed in an Escherichia coli-based cell-free in vitro transcription/translation system, and the crude reactions containing expressed proteins are printed directly onto nitrocellulose microarrays without purification. The protein microarrays are useful for determining the complete antigen-specific humoral immune-response profile from vaccinated or infected humans and animals. The system was verified by cloning, expressing, and printing a vaccinia virus proteome consisting of 185 individual viral proteins. The chips were used to determine Ab profiles in serum from vaccinia virus-immunized humans, primates, and mice. Human serum has high titers of anti-E. coli Abs that require blocking to unmask vaccinia-specific responses. Naive humans exhibit reactivity against a subset of 13 antigens that were not associated with vaccinia immunization. Naive mice and primates lacked this background reactivity. The specific profiles between the three species differed, although a common subset of antigens was reactive after vaccinia immunization. These results verify this platform as a rapid way to comprehensively scan humoral immunity from vaccinated or infected humans and animals.
Figures


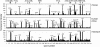
References
-
- Pizza, M., Scarlato, V., Masignani, V., Giuliani, M. M., Arico, B., Comanducci, M., Jennings, G. T., Baldi, L., Bartolini, E., Capecchi, B., et al. (2000) Science 287, 1816-1820. - PubMed
-
- Tettelin, H., Saunders, N. J., Heidelberg, J., Jeffries, A. C., Nelson, K. E., Eisen, J. A., Ketchum, K. A., Hood, D. W., Peden, J. F., Dodson, R. J., et al. (2000) Science 287, 1809-1815. - PubMed
-
- Liang, X., Teng, A., Braun, D. M., Felgner, J., Wang, Y., Baker, S. I., Chen, S., Zelphati, O. & Felgner, P. L. (2002) J. Biol. Chem. 277, 3593-3598. - PubMed
-
- Doolan, D. L., Aguiar, J. C., Weiss, W. R., Sette, A., Felgner, P. L., Regis, D. P., Quinones-Casas, P., Yates, J. R., III, Blair, P. L., Richie, T. L., et al. (2003) J. Exp. Biol. 206, 3789-3802. - PubMed
-
- Crotty, S., Felgner, P., Davies, H., Glidewell, J., Villarreal, L. & Ahmed, R. (2003) J. Immunol. 171, 4969-4973. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

