Probing counterion modulated repulsion and attraction between nucleic acid duplexes in solution
- PMID: 15647360
- PMCID: PMC545826
- DOI: 10.1073/pnas.0404448102
Probing counterion modulated repulsion and attraction between nucleic acid duplexes in solution
Abstract
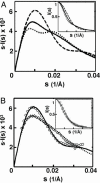
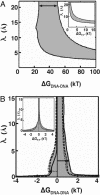
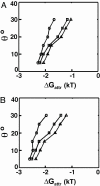
Understanding biological and physical processes involving nucleic acids, such as the binding of proteins to DNA and RNA, DNA condensation, and RNA folding, requires an understanding of the ion atmosphere that surrounds nucleic acids. We have used a simple model DNA system to determine how the ion atmosphere modulates interactions between duplexes in the absence of specific metal ion-binding sites and other complicated interactions. In particular, we have tested whether the Coulomb repulsion between nucleic acids can be reversed by counterions to give a net attraction, as has been proposed recently for the rapid collapse observed early in RNA folding. The conformation of two DNA duplexes tethered by a flexible neutral linker was determined in the presence of a series of cations by small angle x-ray scattering. The small angle x-ray scattering profiles of two control molecules with distinct shapes (a continuous duplex and a mimic of the compact DNA) were in good agreement with predictions, establishing the applicability of this approach. Under low-salt conditions (20 mM Na+), the tethered duplexes are extended because of a Coulombic repulsion estimated to be 2-5 kT/bp. Addition of high concentrations of Na+ (1.2 M), Mg2+ (0.6 M), and spermidine3+ (75 mM) resulted in electrostatic relaxation to a random state. These results indicate that a counterion-induced attractive force between nucleic acid duplexes is not significant under physiological conditions. An upper limit on the magnitude of the attractive potential under all tested ionic conditions is estimated.
Figures
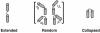
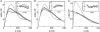
References
-
- Sussman, J. L., Holbrook, S. R., Warrant, R. W., Church, G. M. & Kim, S. H. (1978) J. Mol. Biol. 123, 607–630. - PubMed
-
- Manning, G. S. (1977) Biophys. Chem. 7, 95–102. - PubMed
-
- Manning, G. S. (1978) Biophys. Chem. 9, 65–70. - PubMed
-
- Heilman-Miller, S. L., Thirumalai, D. & Woodson, S. A. (2001) J. Mol. Biol. 306, 1157–1166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

