An Sp1 response element in the Kaposi's sarcoma-associated herpesvirus open reading frame 50 promoter mediates lytic cycle induction by butyrate
- PMID: 15650166
- PMCID: PMC544116
- DOI: 10.1128/JVI.79.3.1397-1408.2005
An Sp1 response element in the Kaposi's sarcoma-associated herpesvirus open reading frame 50 promoter mediates lytic cycle induction by butyrate
Abstract
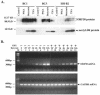
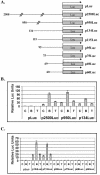
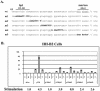
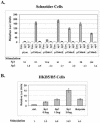
Kaposi's sarcoma-associated herpesvirus (KSHV) can be driven into the lytic cycle in vitro by phorbol esters and sodium butyrate. This report begins to analyze the process by which butyrate activates the promoter of KSHV open reading frame 50 (ORF50), the key viral regulator of the KSHV latency to lytic cycle switch. A short fragment of the promoter, 134 nucleotides upstream of the translational start of ORF50, retained basal uninduced activity and conferred maximal responsiveness to sodium butyrate. The butyrate response element was mapped to a consensus Sp1-binding site. By means of electrophoretic mobility shift assays, both Sp1 and Sp3 were shown to form complexes in vitro with the ORF50 promoter at the Sp1 site. Butyrate induced the formation of a group of novel complexes, including several Sp3-containing complexes, one Sp1-containing complex, and several other complexes that were not identified with antibodies to Sp1 or Sp3. Formation of all butyrate-induced DNA-protein complexes was mediated by the consensus Sp1 site. In insect and mammalian cell lines, Sp1 significantly activated the ORF50 promoter linked to luciferase. Chromatin immunoprecipitation experiments in a PEL cell line showed that butyrate induced Sp1, CBP, and p300 binding to the ORF50 promoter in vivo in an on-off manner. The results suggest that induction of the KSHV lytic cycle by butyrate is mediated through interactions at the Sp1/Sp3 site located 103 to 112 nucleotides upstream of the translational initiation of ORF50 presumably by enhancing the binding of Sp1 to this site.
Figures


Similar articles
-
Sp3 Transcription Factor Cooperates with the Kaposi's Sarcoma-Associated Herpesvirus ORF50 Protein To Synergistically Activate Specific Viral and Cellular Gene Promoters.J Virol. 2020 Aug 31;94(18):e01143-20. doi: 10.1128/JVI.01143-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32641483 Free PMC article.
-
Chromatin remodeling of the Kaposi's sarcoma-associated herpesvirus ORF50 promoter correlates with reactivation from latency.J Virol. 2003 Nov;77(21):11425-35. doi: 10.1128/jvi.77.21.11425-11435.2003. J Virol. 2003. PMID: 14557628 Free PMC article.
-
Positive and negative regulation in the promoter of the ORF46 gene of Kaposi's sarcoma-associated herpesvirus.Virus Res. 2012 May;165(2):157-69. doi: 10.1016/j.virusres.2012.02.010. Epub 2012 Feb 18. Virus Res. 2012. PMID: 22366521
-
Lytic cycle switches of oncogenic human gammaherpesviruses.Adv Cancer Res. 2007;97:81-109. doi: 10.1016/S0065-230X(06)97004-3. Adv Cancer Res. 2007. PMID: 17419942 Review.
-
Kaposi's sarcoma-associated herpesvirus immediate early gene activity.Front Biosci. 2004 Sep 1;9:2245-72. doi: 10.2741/1394. Front Biosci. 2004. PMID: 15353285 Review.
Cited by
-
Molecular Biology of KSHV in Relation to HIV/AIDS-Associated Oncogenesis.Cancer Treat Res. 2019;177:23-62. doi: 10.1007/978-3-030-03502-0_2. Cancer Treat Res. 2019. PMID: 30523620 Free PMC article.
-
Interplay between PKCδ and Sp1 on histone deacetylase inhibitor-mediated Epstein-Barr virus reactivation.J Virol. 2011 Mar;85(5):2373-85. doi: 10.1128/JVI.01602-10. Epub 2010 Dec 15. J Virol. 2011. PMID: 21159880 Free PMC article.
-
Mechanisms of Kaposi's Sarcoma-Associated Herpesvirus Latency and Reactivation.Adv Virol. 2011;2011:193860. doi: 10.1155/2011/193860. Adv Virol. 2011. PMID: 21625290 Free PMC article.
-
Histone deacetylases and the nuclear receptor corepressor regulate lytic-latent switch gene 50 in murine gammaherpesvirus 68-infected macrophages.J Virol. 2010 Nov;84(22):12039-47. doi: 10.1128/JVI.00396-10. Epub 2010 Aug 18. J Virol. 2010. PMID: 20719946 Free PMC article.
-
Histone deacetylase classes I and II regulate Kaposi's sarcoma-associated herpesvirus reactivation.J Virol. 2014 Jan;88(2):1281-92. doi: 10.1128/JVI.02665-13. Epub 2013 Nov 13. J Virol. 2014. PMID: 24227836 Free PMC article.
References
-
- Alkan, S., D. S. Karcher, A. Ortiz, S. Khalil, M. Akhtar, and M. A. Ali. 1997. Human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus in organ transplant patients with immunosuppression. Br. J. Haematol. 96:412-414. - PubMed
-
- Armstrong, S. A., D. A. Barry, R. W. Leggett, and C. R. Mueller. 1997. Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA binding activity. J. Biol. Chem. 272:13489-13495. - PubMed
-
- Ascoli, V., C. M. Mastroianni, V. Galati, M. C. Sirianni, A. Fruscalzo, A. Pistilli, and C. F. Lo. 1998. Primary effusion lymphoma containing human herpesvirus 8 DNA in two AIDS patients with Kaposi's sarcoma. Haematologica 83:8-12. - PubMed
-
- Benasciutti, E., G. Pages, O. Kenzior, W. Folk, F. Blasi, and M. P. Crippa. 2004. MAPK and JNK transduction pathways can phosphorylate Sp1 to activate the uPA minimal promoter elements and endogenous gene transcription. Blood 104:256-262. - PubMed
-
- Bouwman, P., and S. Philipsen. 2002. Regulation of the activity of Sp1-related transcription factors. Mol. Cell. Endocrinol. 195:27-38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

