Repair of the tRNA-like CCA sequence in a multipartite positive-strand RNA virus
- PMID: 15650168
- PMCID: PMC544147
- DOI: 10.1128/JVI.79.3.1417-1427.2005
Repair of the tRNA-like CCA sequence in a multipartite positive-strand RNA virus
Abstract
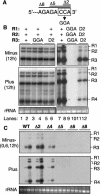
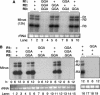
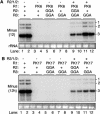
The 3' portions of plus-strand brome mosaic virus (BMV) RNAs mimic cellular tRNAs. Nucleotide substitutions or deletions in the 3'CCA of the tRNA-like sequence (TLS) affect minus-strand initiation unless repaired. We observed that 2-nucleotide deletions involving the CCA 3' sequence in one or all BMV RNAs still allowed RNA accumulation in barley protoplasts at significant levels. Alterations of CCA to GGA in only BMV RNA3 also allowed RNA accumulation at wild-type levels. However, substitutions in all three BMV RNAs severely reduced RNA accumulation, demonstrating that substitutions have different repair requirements than do small deletions. Furthermore, wild-type BMV RNA1 was required for the repair and replication of RNAs with nucleotide substitutions. Results from sequencing of progeny viral RNA from mutant input RNAs demonstrated that RNA1 did not contribute its sequence to the mutant RNAs. Instead, the repaired ends were heterogeneous, with one-third having a restored CCA and others having sequences with the only commonality being the restoration of one cytidylate. The role of BMV RNA1 in increased repair was examined.
Figures



Similar articles
-
Replicase-binding sites on plus- and minus-strand brome mosaic virus RNAs and their roles in RNA replication in plant cells.J Virol. 2004 Dec;78(24):13420-9. doi: 10.1128/JVI.78.24.13420-13429.2004. J Virol. 2004. PMID: 15564452 Free PMC article.
-
Brome mosaic virus RNA syntheses in vitro and in barley protoplasts.J Virol. 2003 May;77(10):5703-11. doi: 10.1128/jvi.77.10.5703-5711.2003. J Virol. 2003. PMID: 12719563 Free PMC article.
-
Sequences 5' of the conserved tRNA-like promoter modulate the initiation of minus-strand synthesis by the brome mosaic virus RNA-dependent RNA polymerase.Virology. 1998 Dec 20;252(2):458-67. doi: 10.1006/viro.1998.9473. Virology. 1998. PMID: 9878626
-
The brome mosaic virus 3' untranslated sequence regulates RNA replication, recombination, and virion assembly.Virus Res. 2015 Aug 3;206:46-52. doi: 10.1016/j.virusres.2015.02.007. Epub 2015 Feb 14. Virus Res. 2015. PMID: 25687214 Review.
-
Non-canonical substrates of aminoacyl-tRNA synthetases: the tRNA-like structure of brome mosaic virus genomic RNA.Biochimie. 1993;75(12):1143-57. doi: 10.1016/0300-9084(93)90014-j. Biochimie. 1993. PMID: 8199250 Review.
Cited by
-
Viral AlkB proteins repair RNA damage by oxidative demethylation.Nucleic Acids Res. 2008 Oct;36(17):5451-61. doi: 10.1093/nar/gkn519. Epub 2008 Aug 21. Nucleic Acids Res. 2008. PMID: 18718927 Free PMC article.
-
Selective repression of translation by the brome mosaic virus 1a RNA replication protein.J Virol. 2007 Feb;81(4):1601-9. doi: 10.1128/JVI.01991-06. Epub 2006 Nov 15. J Virol. 2007. PMID: 17108036 Free PMC article.
-
Role of RNase MRP in viral RNA degradation and RNA recombination.J Virol. 2011 Jan;85(1):243-53. doi: 10.1128/JVI.01749-10. Epub 2010 Oct 20. J Virol. 2011. PMID: 20962095 Free PMC article.
-
Interaction between Brome mosaic virus proteins and RNAs: effects on RNA replication, protein expression, and RNA stability.J Virol. 2005 Nov;79(22):14222-34. doi: 10.1128/JVI.79.22.14222-14234.2005. J Virol. 2005. PMID: 16254357 Free PMC article.
-
Insights into the single-cell reproduction cycle of members of the family Bromoviridae: lessons from the use of protoplast systems.J Virol. 2008 Nov;82(21):10330-40. doi: 10.1128/JVI.00746-08. Epub 2008 Aug 6. J Virol. 2008. PMID: 18684833 Free PMC article. Review. No abstract available.
References
-
- Bacher-Reuven, N. Z. Zhou, and M. P. Deutscher. 1997. Functional overlap of tRNA nucleotidyltrasnferase, poly(A) polymerase I and polynucleotide phosphorylase. J. Biol. Chem. 272:33255-33259. - PubMed
-
- Barrera, I., D. Schuppli, J. M. Sogo, and H. Weber. 1993. Different mechanisms of recognition of bacteriophage Qβ plus and minus strand RNAs by Qβ replicase. J. Mol. Biol. 232:512-521. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

