Sindbis virus translation is inhibited by a PKR/RNase L-independent effector induced by alpha/beta interferon priming of dendritic cells
- PMID: 15650175
- PMCID: PMC544143
- DOI: 10.1128/JVI.79.3.1487-1499.2005
Sindbis virus translation is inhibited by a PKR/RNase L-independent effector induced by alpha/beta interferon priming of dendritic cells
Abstract
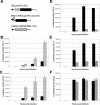
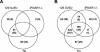
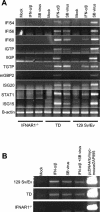
The tropism of Sindbis virus (SB) for cells of the dendritic cell (DC) lineage and the virulence of SB in vivo are largely determined by the efficacy of alpha/beta interferon (IFN-alpha/beta)-mediated antiviral responses. These responses are essentially intact in the absence of PKR and/or RNase L (K. D. Ryman, L. J. White, R. E. Johnston, and W. B. Klimstra, Viral Immunol. 15:53-76, 2002). In the present studies, we investigated the nature of antiviral effects and identity of antiviral effectors primed by IFN-alpha/beta treatment of bone marrow-derived DCs (BMDCs) generated from mice deficient in PKR and RNase L (TD). IFN-alpha/beta priming exerted significant antiviral activity at very early stages of SB replication and most likely inhibited the initial translation of infecting genomes. The early effect targeted cap-dependent translation as protein synthesis from an SB-like and a simple RNA were inhibited by interferon treatment, but an encephalomyocarditis virus internal ribosome entry site-driven element exhibited no inhibition. Phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2 was defective after virus infection of TD cells, suggesting other mechanisms of translation inhibition. To identify components of these alternative antiviral pathway(s), we have compared global gene regulation in BMDCs derived from normal 129 Sv/Ev, IFNAR1-/-, and TD mice following infection with SB or treatment with IFN-alpha/beta. Candidate effectors of alternative antiviral pathways were those genes induced by virus infection or IFN-alpha/beta treatment in 129 Sv/Ev and TD-derived BMDC but not in virus-infected or IFN-alpha/beta-treated IFNAR1-/- cells. Statistical analyses of gene array data identified 44 genes that met these criteria which are discussed.
Figures



References
-
- Basler, C. F., and A. Garcia-Sastre. 2002. Viruses and the type I interferon antiviral system: induction and evasion. Int. Rev. Immunol. 21:305-337. - PubMed
-
- Burgess, T. H., K. E. Steele, B. A. Schoneboom, and F. B. Grieder. 2001. Clinicopathologic features of viral agents of potential use by bioterrorists. Clin. Lab. Med. 21:475-493. - PubMed
-
- David, M., E. Petricoin III, C. Benjamin, R. Pine, M. J. Weber, and A. C. Larner. 1995. Requirement for MAP kinase (ERK2) activity in interferon alpha- and interferon beta-stimulated gene expression through STAT proteins. Science 269:1721-1723. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

