Complex formation between the UL16 and UL21 tegument proteins of pseudorabies virus
- PMID: 15650177
- PMCID: PMC544144
- DOI: 10.1128/JVI.79.3.1510-1522.2005
Complex formation between the UL16 and UL21 tegument proteins of pseudorabies virus
Abstract
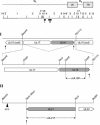
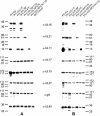
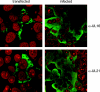
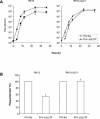
The products of the UL16 and UL21 genes represent tegument proteins which are conserved throughout the mammalian herpesviruses. To identify and functionally characterize the respective proteins in the alphaherpesvirus pseudorabies virus, monospecific antisera against bacterially expressed fusion proteins were generated. In immunoblots the UL16 antiserum detected a ca. 40-kDa protein in infected cells and purified virion preparations, whereas the anti-UL21 serum recognized a protein of approximately 60 kDa. Interestingly, in immunoprecipitations using either antiserum, both proteins were coprecipitated, demonstrating the formation of a physical complex. To investigate protein function, viruses lacking either UL16, UL21, or both were constructed. Mutant viruses could be propagated on noncomplementing cells, indicating that these proteins, either alone or in combination, are not required for viral replication in cell culture. However, plaque sizes and viral titers were reduced. Electron microscopy showed only slight alterations in cytoplasmic virion morphogenesis, whereas intranuclear maturation stages were not affected. Similar results were obtained with a triple mutant simultaneously lacking the three conserved tegument proteins UL11, UL16, and UL21. In summary, our results uncover a novel interaction between conserved herpesvirus tegument proteins that increases the complexity of the intricate network of protein-protein interactions involved in herpesvirus morphogenesis.
Figures




Similar articles
-
Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm.J Virol. 2002 Sep;76(17):8820-33. doi: 10.1128/jvi.76.17.8820-8833.2002. J Virol. 2002. PMID: 12163602 Free PMC article.
-
The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions.J Virol. 2002 Jul;76(13):6729-42. doi: 10.1128/jvi.76.13.6729-6742.2002. J Virol. 2002. PMID: 12050386 Free PMC article.
-
The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm.J Virol. 2003 May;77(9):5339-51. doi: 10.1128/jvi.77.9.5339-5351.2003. J Virol. 2003. PMID: 12692236 Free PMC article.
-
Functional analysis of the pseudorabies virus UL51 protein.J Virol. 2005 Mar;79(6):3831-40. doi: 10.1128/JVI.79.6.3831-3840.2005. J Virol. 2005. PMID: 15731276 Free PMC article.
-
Features and Functions of the Conserved Herpesvirus Tegument Protein UL11 and Its Binding Partners.Front Microbiol. 2022 Jun 3;13:829754. doi: 10.3389/fmicb.2022.829754. eCollection 2022. Front Microbiol. 2022. PMID: 35722336 Free PMC article. Review.
Cited by
-
Novel Structure and Unexpected RNA-Binding Ability of the C-Terminal Domain of Herpes Simplex Virus 1 Tegument Protein UL21.J Virol. 2016 May 27;90(12):5759-69. doi: 10.1128/JVI.00475-16. Print 2016 Jun 15. J Virol. 2016. PMID: 27053559 Free PMC article.
-
Conserved Outer Tegument Component UL11 from Herpes Simplex Virus 1 Is an Intrinsically Disordered, RNA-Binding Protein.mBio. 2020 May 5;11(3):e00810-20. doi: 10.1128/mBio.00810-20. mBio. 2020. PMID: 32371601 Free PMC article.
-
Effects of simultaneous deletion of pUL11 and glycoprotein M on virion maturation of herpes simplex virus type 1.J Virol. 2009 Jan;83(2):896-907. doi: 10.1128/JVI.01842-08. Epub 2008 Nov 12. J Virol. 2009. PMID: 19004941 Free PMC article.
-
Assembly and Egress of an Alphaherpesvirus Clockwork.Adv Anat Embryol Cell Biol. 2017;223:171-193. doi: 10.1007/978-3-319-53168-7_8. Adv Anat Embryol Cell Biol. 2017. PMID: 28528444 Free PMC article. Review.
-
pUL21 is a viral phosphatase adaptor that promotes herpes simplex virus replication and spread.PLoS Pathog. 2021 Aug 16;17(8):e1009824. doi: 10.1371/journal.ppat.1009824. eCollection 2021 Aug. PLoS Pathog. 2021. PMID: 34398933 Free PMC article.
References
-
- Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. F. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. Tuffnell, and B. G. Barrell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

