Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein
- PMID: 15650187
- PMCID: PMC544121
- DOI: 10.1128/JVI.79.3.1613-1622.2005
Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein
Abstract
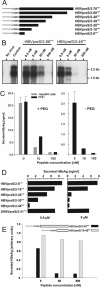
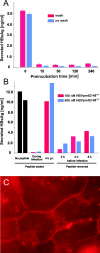
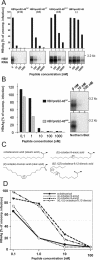
The lack of an appropriate in vitro infection system for the major human pathogen hepatitis B virus (HBV) has prevented a molecular understanding of the early infection events of HBV. We used the novel HBV-infectible cell line HepaRG and primary human hepatocytes to investigate the interference of infection by HBV envelope protein-derived peptides. We found that a peptide consisting of the authentically myristoylated N-terminal 47 amino acids of the pre-S1 domain of the large viral envelope protein (L protein) specifically prevented HBV infection, with a 50% inhibitory concentration (IC50) of 8 nM. The replacement of myristic acid with other hydrophobic moieties resulted in changes in the inhibitory activity, most notably by a decrease in the IC50 to picomolar concentrations for longer unbranched fatty acids. The obstruction of HepaRG cell susceptibility to HBV infection after short preincubation times with the peptides suggested that the peptides efficiently target and inactivate a receptor at the hepatocyte surface. Our data both shed light on the molecular mechanism of HBV entry into hepatocytes and provide a basis for the development of potent hepadnaviral entry inhibitors as a novel therapeutic concept for the treatment of hepatitis Beta.
Figures


Similar articles
-
Characterization of a hepatitis B and hepatitis delta virus receptor binding site.Hepatology. 2006 Apr;43(4):750-60. doi: 10.1002/hep.21112. Hepatology. 2006. PMID: 16557545
-
N-terminal myristoylation-dependent masking of neutralizing epitopes in the preS1 attachment site of hepatitis B virus.J Hepatol. 2011 Jul;55(1):29-37. doi: 10.1016/j.jhep.2010.10.019. Epub 2010 Nov 28. J Hepatol. 2011. PMID: 21145866
-
Myristylation of the large surface protein is required for hepatitis B virus in vitro infectivity.Virology. 1996 Apr 15;218(2):396-9. doi: 10.1006/viro.1996.0209. Virology. 1996. PMID: 8610467
-
Entry of hepatitis B virus: mechanism and new therapeutic target.Pathol Biol (Paris). 2010 Aug;58(4):301-7. doi: 10.1016/j.patbio.2010.04.001. Epub 2010 May 31. Pathol Biol (Paris). 2010. PMID: 20570056 Review.
-
Attachment sites and neutralising epitopes of hepatitis B virus.Minerva Gastroenterol Dietol. 2006 Mar;52(1):3-21. Minerva Gastroenterol Dietol. 2006. PMID: 16554703 Review.
Cited by
-
Advances with RNAi-Based Therapy for Hepatitis B Virus Infection.Viruses. 2020 Aug 4;12(8):851. doi: 10.3390/v12080851. Viruses. 2020. PMID: 32759756 Free PMC article. Review.
-
Advances in HBV infection and replication systems in vitro.Virol J. 2021 May 29;18(1):105. doi: 10.1186/s12985-021-01580-6. Virol J. 2021. PMID: 34051803 Free PMC article. Review.
-
Entry of hepatitis B and hepatitis D virus into hepatocytes: Basic insights and clinical implications.J Hepatol. 2016 Apr;64(1 Suppl):S32-S40. doi: 10.1016/j.jhep.2016.02.011. J Hepatol. 2016. PMID: 27084034 Free PMC article. Review.
-
Structural basis of hepatitis B virus receptor binding.Nat Struct Mol Biol. 2024 Mar;31(3):447-454. doi: 10.1038/s41594-023-01191-5. Epub 2024 Jan 17. Nat Struct Mol Biol. 2024. PMID: 38233573
-
Troglitazone Impedes the Oligomerization of Sodium Taurocholate Cotransporting Polypeptide and Entry of Hepatitis B Virus Into Hepatocytes.Front Microbiol. 2019 Jan 8;9:3257. doi: 10.3389/fmicb.2018.03257. eCollection 2018. Front Microbiol. 2019. PMID: 30671048 Free PMC article.
References
-
- Berting, A., C. Fischer, S. Schaefer, W. Garten, H. D. Klenk, and W. H. Gerlich. 2000. Hemifusion activity of a chimeric influenza virus hemagglutinin with a putative fusion peptide from hepatitis B virus. Virus Res. 68:35-49. - PubMed
-
- Bruss, V., J. Hagelstein, E. Gerhardt, and P. R. Galle. 1996. Myristylation of the large surface protein is required for hepatitis B virus in vitro infectivity. Virology 218:396-399. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

