Computational design of antiviral RNA interference strategies that resist human immunodeficiency virus escape
- PMID: 15650190
- PMCID: PMC544124
- DOI: 10.1128/JVI.79.3.1645-1654.2005
Computational design of antiviral RNA interference strategies that resist human immunodeficiency virus escape
Abstract
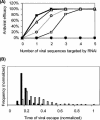
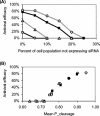
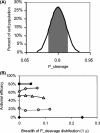
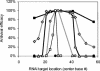
Recently developed antiviral strategies based upon RNA interference (RNAi), which harnesses an innate cellular system for the targeted down-regulation of gene expression, appear highly promising and offer alternative approaches to conventional highly active antiretroviral therapy or efforts to develop an AIDS vaccine. However, RNAi is faced with several challenges that must be overcome to fully realize its promise. Specifically, it degrades target RNA in a highly sequence-specific manner and is thus susceptible to viral mutational escape, and there are also challenges in delivery systems to induce RNAi. To aid in the development of anti-human immunodeficiency virus (anti-HIV) RNAi therapies, we have developed a novel stochastic computational model that simulates in molecular-level detail the propagation of an HIV infection in cells expressing RNAi. The model provides quantitative predictions on how targeting multiple locations in the HIV genome, while keeping the overall RNAi strength constant, significantly improves efficacy. Furthermore, it demonstrates that delivery systems must be highly efficient to preclude leaving reservoirs of unprotected cells where the virus can propagate, mutate, and eventually overwhelm the entire system. It also predicts how therapeutic success depends upon a relationship between RNAi strength and delivery efficiency and uniformity. Finally, targeting an essential viral element, in this case the HIV TAR region, can be highly successful if the RNAi target sequence is correctly selected. In addition to providing specific predictions for how to optimize a clinical therapy, this system may also serve as a future tool for investigating more fundamental questions of viral evolution.
Figures

Similar articles
-
Control of HIV-1 replication by RNA interference.Virus Res. 2004 Jun 1;102(1):53-8. doi: 10.1016/j.virusres.2004.01.015. Virus Res. 2004. PMID: 15068880 Review.
-
Silencing of HIV-1 with RNA interference: a multiple shRNA approach.Mol Ther. 2006 Dec;14(6):883-92. doi: 10.1016/j.ymthe.2006.07.007. Epub 2006 Sep 7. Mol Ther. 2006. PMID: 16959541
-
HIV-1-specific RNA interference.Curr Opin Mol Ther. 2004 Aug;6(4):373-80. Curr Opin Mol Ther. 2004. PMID: 15468596 Review.
-
RNAi in combination with a ribozyme and TAR decoy for treatment of HIV infection in hematopoietic cell gene therapy.Ann N Y Acad Sci. 2006 Oct;1082:172-9. doi: 10.1196/annals.1348.006. Ann N Y Acad Sci. 2006. PMID: 17145937 Review.
-
RNA interference and HIV-1 infection.Rev Med Virol. 2008 Jan-Feb;18(1):5-18. doi: 10.1002/rmv.553. Rev Med Virol. 2008. PMID: 17764099 Review.
Cited by
-
Cassette deletion in multiple shRNA lentiviral vectors for HIV-1 and its impact on treatment success.Virol J. 2009 Oct 30;6:184. doi: 10.1186/1743-422X-6-184. Virol J. 2009. PMID: 19878571 Free PMC article.
-
Kinetic Modeling of Virus Growth in Cells.Microbiol Mol Biol Rev. 2018 Mar 28;82(2):e00066-17. doi: 10.1128/MMBR.00066-17. Print 2018 Jun. Microbiol Mol Biol Rev. 2018. PMID: 29592895 Free PMC article. Review.
-
The anti-genomic (negative) strand of Hepatitis C Virus is not targetable by shRNA.Nucleic Acids Res. 2013 Apr 1;41(6):3688-98. doi: 10.1093/nar/gkt068. Epub 2013 Feb 8. Nucleic Acids Res. 2013. PMID: 23396439 Free PMC article.
-
Preclinical evaluation of an anti-HCV miRNA cluster for treatment of HCV infection.Mol Ther. 2013 Mar;21(3):588-601. doi: 10.1038/mt.2012.247. Epub 2013 Jan 8. Mol Ther. 2013. PMID: 23295950 Free PMC article.
-
Combinatorial RNAi: a winning strategy for the race against evolving targets?Mol Ther. 2007 May;15(5):878-88. doi: 10.1038/sj.mt.6300116. Epub 2007 Feb 20. Mol Ther. 2007. PMID: 17311009 Free PMC article. Review.
References
-
- Anderson, J., A. Banerjea, and R. Akkina. 2003. Bispecific short hairpin siRNA constructs targeted to CD4, CXCR4, and CCR5 confer HIV-1 resistance. Oligonucleotides 13:303-312. - PubMed
-
- Banerjea, A., M. J. Li, G. Bauer, L. Remling, N. S. Lee, J. Rossi, and R. Akkina. 2003. Inhibition of HIV-1 by lentiviral vector-transduced siRNAs in T lymphocytes differentiated in SCID-hu mice and CD34+ progenitor cell-derived macrophages. Mol. Ther. 8:62-71. - PubMed
-
- Berkhout, B. 2004. RNA interference as an antiviral approach: targeting HIV-1. Curr. Opin. Mol. Ther. 6:141-145. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

