Probing a rate-limiting step by mutational perturbation of AdoMet binding in the HhaI methyltransferase
- PMID: 15653631
- PMCID: PMC546160
- DOI: 10.1093/nar/gki175
Probing a rate-limiting step by mutational perturbation of AdoMet binding in the HhaI methyltransferase
Abstract
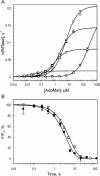
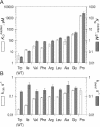
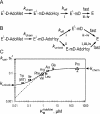
DNA methylation plays important roles via regulation of numerous cellular mechanisms in diverse organisms, including humans. The paradigm bacterial methyltransferase (MTase) HhaI (M.HhaI) catalyzes the transfer of a methyl group from the cofactor S-adenosyl-L-methionine (AdoMet) onto the target cytosine in DNA, yielding 5-methylcytosine and S-adenosyl-L-homocysteine (AdoHcy). The turnover rate (k cat) of M.HhaI, and the other two cytosine-5 MTases examined, is limited by a step subsequent to methyl transfer; however, no such step has so far been identified. To elucidate the role of cofactor interactions during catalysis, eight mutants of Trp41, which is located in the cofactor binding pocket, were constructed and characterized. The mutants show full proficiency in DNA binding and base-flipping, and little variation is observed in the apparent methyl transfer rate k chem as determined by rapid-quench experiments using immobilized fluorescent-labeled DNA. However, the Trp41 replacements with short side chains substantially perturb cofactor binding (100-fold higher K(AdoMet)D and K(AdoMet)M) leading to a faster turnover of the enzyme (10-fold higher k cat). Our analysis indicates that the rate-limiting breakdown of a long-lived ternary product complex is initiated by the dissociation of AdoHcy or the opening of the catalytic loop in the enzyme.
Figures


Similar articles
-
S-adenosyl-L-methionine-dependent methyl transfer: observable precatalytic intermediates during DNA cytosine methylation.Biochemistry. 2007 Jul 31;46(30):8766-75. doi: 10.1021/bi7005948. Epub 2007 Jul 7. Biochemistry. 2007. PMID: 17616174
-
The mechanism of DNA cytosine-5 methylation. Kinetic and mutational dissection of Hhai methyltransferase.J Biol Chem. 2001 Jun 15;276(24):20924-34. doi: 10.1074/jbc.M101429200. Epub 2001 Mar 29. J Biol Chem. 2001. PMID: 11283006
-
The role of Arg165 towards base flipping, base stabilization and catalysis in M.HhaI.J Mol Biol. 2006 Sep 22;362(3):516-27. doi: 10.1016/j.jmb.2006.07.030. Epub 2006 Jul 22. J Mol Biol. 2006. PMID: 16926025
-
Structure, function, and mechanism of HhaI DNA methyltransferases.Crit Rev Biochem Mol Biol. 2002;37(3):167-97. doi: 10.1080/10409230290771492. Crit Rev Biochem Mol Biol. 2002. PMID: 12139442 Review.
-
DNA methyltransferases: mechanistic models derived from kinetic analysis.Crit Rev Biochem Mol Biol. 2012 Mar-Apr;47(2):97-193. doi: 10.3109/10409238.2011.620942. Epub 2012 Jan 20. Crit Rev Biochem Mol Biol. 2012. PMID: 22260147 Review.
Cited by
-
Metadynamics simulation study on the conformational transformation of HhaI methyltransferase: an induced-fit base-flipping hypothesis.Biomed Res Int. 2014;2014:304563. doi: 10.1155/2014/304563. Epub 2014 Jun 19. Biomed Res Int. 2014. PMID: 25045662 Free PMC article.
-
Inactive DNMT3B splice variants modulate de novo DNA methylation.PLoS One. 2013 Jul 19;8(7):e69486. doi: 10.1371/journal.pone.0069486. Print 2013. PLoS One. 2013. PMID: 23894490 Free PMC article.
-
Engineering the DNA cytosine-5 methyltransferase reaction for sequence-specific labeling of DNA.Nucleic Acids Res. 2012 Dec;40(22):11594-602. doi: 10.1093/nar/gks914. Epub 2012 Oct 5. Nucleic Acids Res. 2012. PMID: 23042683 Free PMC article.
-
Coupling sequence-specific recognition to DNA modification.J Biol Chem. 2009 Aug 21;284(34):22690-6. doi: 10.1074/jbc.M109.015966. Epub 2009 Jun 4. J Biol Chem. 2009. PMID: 19497854 Free PMC article.
-
A directed evolution design of a GCG-specific DNA hemimethylase.Nucleic Acids Res. 2009 Nov;37(21):7332-41. doi: 10.1093/nar/gkp772. Nucleic Acids Res. 2009. PMID: 19783820 Free PMC article.
References
-
- Robertson K.D., Jones P.A. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21:461–467. - PubMed
-
- Cheng X., Kumar S., Posfai J., Pflugrath J.W., Roberts R.J. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-l-methionine. Cell. 1993;74:299–307. - PubMed
-
- Klimasauskas S., Kumar S., Roberts R.J., Cheng X. Hhal methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

