Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins
- PMID: 15653639
- PMCID: PMC546159
- DOI: 10.1093/nar/gki174
Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins
Abstract
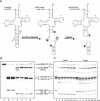
Pre-tRNA splicing is an essential process in all eukaryotes. It requires the concerted action of an endonuclease to remove the intron and a ligase for joining the resulting tRNA halves as studied best in the yeast Saccharomyces cerevisiae. Here, we report the first characterization of an RNA ligase protein and its gene from a higher eukaryotic organism that is an essential component of the pre-tRNA splicing process. Purification of tRNA ligase from wheat germ by successive column chromatographic steps has identified a protein of 125 kDa by its potentiality to covalently bind AMP, and by its ability to catalyse the ligation of tRNA halves and the circularization of linear introns. Peptide sequences obtained from the purified protein led to the elucidation of the corresponding proteins and their genes in Arabidopsis and Oryza databases. The plant tRNA ligases exhibit no overall sequence homologies to any known RNA ligases, however, they harbour a number of conserved motifs that indicate the presence of three intrinsic enzyme activities: an adenylyltransferase/ligase domain in the N-terminal region, a polynucleotide kinase in the centre and a cyclic phosphodiesterase domain at the C-terminal end. In vitro expression of the recombinant Arabidopsis tRNA ligase and functional analyses revealed all expected individual activities. Plant RNA ligases are active on a variety of substrates in vitro and are capable of inter- and intramolecular RNA joining. Hence, we conclude that their role in vivo might comprise yet unknown essential functions besides their involvement in pre-tRNA splicing.
Figures





References
-
- Uhlenbeck O.C. T4 RNA ligase. Trends Biochem. Sci. 1983;8:94–96.
-
- Heaphy S., Singh M., Gait M.J. Effect of single amino acid changes in the region of the adenylylation site of T4 RNA ligase. Biochemistry. 1987;26:1688–1696. - PubMed
-
- Wang L.K., Ho C.K., Pei Y., Shuman S. Mutational analysis of bacteriophage T4 RNA ligase 1. Different functional groups are required for the nucleotidyl transfer and phosphodiester bond formation of the ligation reaction. J. Biol. Chem. 2003;278:29454–29462. - PubMed
-
- Kaufmann G. Anticodon nucleases. Trends Biochem. Sci. 2000;25:70–74. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

