Probing the DNA kink structure induced by the hyperthermophilic chromosomal protein Sac7d
- PMID: 15653643
- PMCID: PMC546169
- DOI: 10.1093/nar/gki191
Probing the DNA kink structure induced by the hyperthermophilic chromosomal protein Sac7d
Abstract
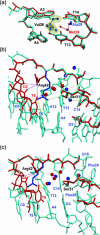
Sac7d, a small, abundant, sequence-general DNA-binding protein from the hyperthermophilic archaeon Sulfolobus acidocaldarius, causes a single-step sharp kink in DNA (approximately 60 degrees) via the intercalation of both Val26 and Met29. These two amino acids were systematically changed in size to probe their effects on DNA kinking. Eight crystal structures of five Sac7d mutant-DNA complexes have been analyzed. The DNA-binding pattern of the V26A and M29A single mutants is similar to that of the wild-type, whereas the V26A/M29A protein binds DNA without side chain intercalation, resulting in a smaller overall bending (approximately 50 degrees). The M29F mutant inserts the Phe29 side chain orthogonally to the C2pG3 step without stacking with base pairs, inducing a sharp kink (approximately 80 degrees). In the V26F/M29F-GCGATCGC complex, Phe26 intercalates deeply into DNA bases by stacking with the G3 base, whereas Phe29 is stacked on the G15 deoxyribose, in a way similar to those used by the TATA box-binding proteins. All mutants have reduced DNA-stabilizing ability, as indicated by their lower T m values. The DNA kink patterns caused by different combinations of hydrophobic side chains may be relevant in understanding the manner by which other minor groove-binding proteins interact with DNA.
Figures


Similar articles
-
The hyperthermophile chromosomal protein Sac7d sharply kinks DNA.Nature. 1998 Mar 12;392(6672):202-5. doi: 10.1038/32455. Nature. 1998. PMID: 9515968
-
The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA.Nat Struct Biol. 1998 Sep;5(9):782-6. doi: 10.1038/1822. Nat Struct Biol. 1998. PMID: 9731772
-
Structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers.Acta Crystallogr D Biol Crystallogr. 2004 Aug;60(Pt 8):1381-7. doi: 10.1107/S0907444904012065. Epub 2004 Jul 21. Acta Crystallogr D Biol Crystallogr. 2004. PMID: 15272160
-
Intercalation, DNA kinking, and the control of transcription.Science. 1996 Feb 9;271(5250):778-84. doi: 10.1126/science.271.5250.778. Science. 1996. PMID: 8628992 Review.
-
Nonsequence-specific DNA recognition: a structural perspective.Structure. 2000 Apr 15;8(4):R83-9. doi: 10.1016/s0969-2126(00)00126-x. Structure. 2000. PMID: 10801483 Review.
Cited by
-
Probing the role of intercalating protein sidechains for kink formation in DNA.PLoS One. 2018 Feb 12;13(2):e0192605. doi: 10.1371/journal.pone.0192605. eCollection 2018. PLoS One. 2018. PMID: 29432448 Free PMC article.
-
Twist and turn: a revised structural view on the unpaired bubble of class II CPD photolyase in complex with damaged DNA.IUCrJ. 2018 Aug 8;5(Pt 5):608-618. doi: 10.1107/S205225251800996X. eCollection 2018 Sep 1. IUCrJ. 2018. PMID: 30224964 Free PMC article.
-
What drives proteins into the major or minor grooves of DNA?J Mol Biol. 2007 Jan 5;365(1):1-9. doi: 10.1016/j.jmb.2006.09.059. Epub 2006 Sep 27. J Mol Biol. 2007. PMID: 17055530 Free PMC article. Review.
-
Biochemical and structural characterization of Cren7, a novel chromatin protein conserved among Crenarchaea.Nucleic Acids Res. 2008 Mar;36(4):1129-37. doi: 10.1093/nar/gkm1128. Epub 2007 Dec 20. Nucleic Acids Res. 2008. PMID: 18096617 Free PMC article.
-
Structural analysis of Rtt106p reveals a DNA binding role required for heterochromatin silencing.J Biol Chem. 2010 Feb 5;285(6):4251-4262. doi: 10.1074/jbc.M109.055996. Epub 2009 Dec 10. J Biol Chem. 2010. PMID: 20007951 Free PMC article.
References
-
- Hagerman P.J. Sequence-directed curvature of DNA. Annu. Rev. Biochem. 1990;59:755–781. - PubMed
-
- McFall S.M., Chugani S.A., Chakrabarty M.A. Transcriptional activation of the catechol and chlorocatechol operons: variations on a theme. Gene. 1998;223:257–267. - PubMed
-
- McGill G., Fisher D.E. DNA bending and the curious case of Fos/Jun. Chem. Biol. 1998;5:R29–R38. - PubMed
-
- Zahn K., Blattner F.R. Sequence-induced DNA curvature at the bacteriophage lambda origin of replication. Nature. 1985;317:451–453. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

