Protein disorder: conformational distribution of the flexible linker in a chimeric double cellulase
- PMID: 15653742
- PMCID: PMC1305377
- DOI: 10.1529/biophysj.104.050146
Protein disorder: conformational distribution of the flexible linker in a chimeric double cellulase
Abstract
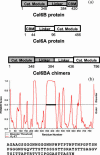
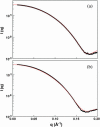
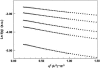
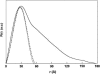
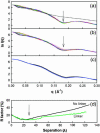
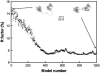
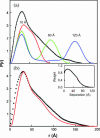
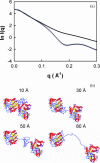
The structural properties of the linker peptide connecting the cellulose-binding module to the catalytic module in bimodular cellulases have been investigated by small-angle x-ray scattering. Since the linker and the cellulose-binding module are relatively small and cannot be readily detected separately, the conformation of the linker was studied by means of an artificial fusion protein, Cel6BA, in which an 88-residue linker connects the large catalytic modules of the cellulases Cel6A and Cel6B from Humicola insolens. Our data showed that Cel6BA is very elongated with a maximum dimension of 178 A, but could not be described by a single conformation. Modeling of a series of Cel6BA conformers with interdomain separations ranging between 10 A and 130 A showed that good Guinier and P(r) profile fits were obtained by a weighted average of the scattering curves of all the models where the linker follows a nonrandom distribution, with a preference for the more compact conformers. These structural properties are likely to be essential for the function of the linker as a molecular spring between the two functional modules. Small-angle x-ray scattering therefore provides a unique tool to quantitatively analyze the conformational disorder typical of proteins described as natively unfolded.
Figures
Similar articles
-
Dimension, shape, and conformational flexibility of a two domain fungal cellulase in solution probed by small angle X-ray scattering.J Biol Chem. 2002 Oct 25;277(43):40887-92. doi: 10.1074/jbc.M205404200. Epub 2002 Aug 16. J Biol Chem. 2002. PMID: 12186865
-
Small angle X-ray scattering analysis of Clostridium thermocellum cellulosome N-terminal complexes reveals a highly dynamic structure.J Biol Chem. 2013 Mar 15;288(11):7978-7985. doi: 10.1074/jbc.M112.408757. Epub 2013 Jan 22. J Biol Chem. 2013. PMID: 23341454 Free PMC article.
-
Structure of a full length psychrophilic cellulase from Pseudoalteromonas haloplanktis revealed by X-ray diffraction and small angle X-ray scattering.J Mol Biol. 2005 May 20;348(5):1211-24. doi: 10.1016/j.jmb.2005.03.026. Epub 2005 Mar 25. J Mol Biol. 2005. PMID: 15854656
-
Cellulose, cellulases and cellulosomes.Curr Opin Struct Biol. 1998 Oct;8(5):548-57. doi: 10.1016/s0959-440x(98)80143-7. Curr Opin Struct Biol. 1998. PMID: 9818257 Review.
-
Structure and function analysis of Pseudomonas plant cell wall hydrolases.Prog Nucleic Acid Res Mol Biol. 1998;61:211-41. doi: 10.1016/s0079-6603(08)60828-4. Prog Nucleic Acid Res Mol Biol. 1998. PMID: 9752722 Review.
Cited by
-
Intrinsically Disordered Linkers Impart Processivity on Enzymes by Spatial Confinement of Binding Domains.Int J Mol Sci. 2019 Apr 29;20(9):2119. doi: 10.3390/ijms20092119. Int J Mol Sci. 2019. PMID: 31032817 Free PMC article.
-
The O-glycosylated linker from the Trichoderma reesei Family 7 cellulase is a flexible, disordered protein.Biophys J. 2010 Dec 1;99(11):3773-81. doi: 10.1016/j.bpj.2010.10.032. Biophys J. 2010. PMID: 21112302 Free PMC article.
-
Understanding the structural ensembles of a highly extended disordered protein.Mol Biosyst. 2012 Jan;8(1):308-19. doi: 10.1039/c1mb05243h. Epub 2011 Oct 6. Mol Biosyst. 2012. PMID: 21979461 Free PMC article.
-
Structure and flexibility within proteins as identified through small angle X-ray scattering.Gen Physiol Biophys. 2009 Jun;28(2):174-89. doi: 10.4149/gpb_2009_02_174. Gen Physiol Biophys. 2009. PMID: 19592714 Free PMC article.
-
How random are intrinsically disordered proteins? A small angle scattering perspective.Curr Protein Pept Sci. 2012 Feb;13(1):55-75. doi: 10.2174/138920312799277901. Curr Protein Pept Sci. 2012. PMID: 22044150 Free PMC article. Review.
References
-
- Ashton, A. W., M. K. Boehm, J. R. Gallimore, M. B. Pepys, and S. J. Perkins. 1997. Pentameric and decameric structures in solution of serum amyloid P component by x-ray and neutron scattering and molecular modelling analyses. J. Mol. Biol. 272:408–422. - PubMed
-
- Beavil, A. J., R. J. Young, B. J. Sutton, and S. J. Perkins. 1995. Bent domain structure of recombinant human IgE-Fc in solution by x-ray and neutron scattering in conjunction with an automated curve-fitting procedure. Biochemistry. 34:14449–14461. - PubMed
-
- Bergmann, A., G. Fritz, and O. Glatter. 2000. Solving the generalized indirect Fourier transformation (GIFT) by Boltzmann simplex simulated annealing (BSSA). J. Appl. Crystallogr. 33:1212–1216.
-
- Boehm, M. K., J. M. Woof, M. A. Kerr, and S. J. Perkins. 1999. The Fab and Fc fragments of IgA1 exhibit a different arrangement from that in IgG: a study by x-ray and neutron solution scattering and homology modelling. J. Mol. Biol. 286:1421–1447. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

