Characterization of potential selenium-binding proteins in the selenophosphate synthetase system
- PMID: 15653770
- PMCID: PMC545862
- DOI: 10.1073/pnas.0409042102
Characterization of potential selenium-binding proteins in the selenophosphate synthetase system
Abstract
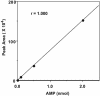
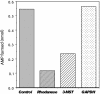
Selenophosphate, an activated form of selenium that can serve as a selenium donor, is generated by the selD gene product, selenophosphate synthetase (SPS). Selenophosphate is required by several bacteria and by mammals for the specific synthesis of Secys-tRNA, the precursor of selenocysteine in selenoenzymes. Although free selenide can be used in vitro for synthesis of selenophosphate, the physiological system that donates selenium to SPS is incompletely characterized. To detect potential selenium-delivery proteins, two known sulfurtransferases and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; EC 1.2.1.12) were analyzed for ability to bind and transfer selenium. Rhodanese (EC 2.8.1.1) was shown to bind selenium tightly, with only part of the selenium being available as substrate for SPS in the presence of added reductant. 3-Mercaptopyruvate sulfurtransferase (3-MST; EC 2.8.1.2) and GAPDH also bound selenium supplied as selenodiglutathione formed from SeO3(2-) and glutathione. Selenium bound to 3-MST and GAPDH was released more readily than that from rhodanese and also was more available as a substrate for SPS. Although rhodanese retained tightly bound selenium under aerobic conditions, the protein gradually became insoluble, whereas GAPDH containing bound selenium was stable at neutral pH for a long period. These results indicate that 3-MST and GAPDH have more suitable potentials as a physiological selenium-delivery protein than rhodanese. In the presence of a selenium-binding protein, a low level of selenodiglutathione formed from SeO3(2-) and glutathione could effectively replace the high concentrations of selenide routinely used as substrate in the SPS in vitro assays.
Figures



Similar articles
-
Formation of a selenium-substituted rhodanese by reaction with selenite and glutathione: possible role of a protein perselenide in a selenium delivery system.Proc Natl Acad Sci U S A. 2001 Aug 14;98(17):9494-8. doi: 10.1073/pnas.171320998. Epub 2001 Aug 7. Proc Natl Acad Sci U S A. 2001. PMID: 11493708 Free PMC article.
-
Delivery of selenium to selenophosphate synthetase for selenoprotein biosynthesis.Biochim Biophys Acta Gen Subj. 2018 Nov;1862(11):2433-2440. doi: 10.1016/j.bbagen.2018.05.023. Epub 2018 Jun 1. Biochim Biophys Acta Gen Subj. 2018. PMID: 29859962 Review.
-
Escherichia coli NifS-like proteins provide selenium in the pathway for the biosynthesis of selenophosphate.J Biol Chem. 2000 Aug 4;275(31):23769-73. doi: 10.1074/jbc.M000926200. J Biol Chem. 2000. PMID: 10829016
-
Cloning and functional characterization of human selenophosphate synthetase, an essential component of selenoprotein synthesis.J Biol Chem. 1995 Sep 15;270(37):21659-64. doi: 10.1074/jbc.270.37.21659. J Biol Chem. 1995. PMID: 7665581
-
Utilization of selenocysteine as a source of selenium for selenophosphate biosynthesis.Biofactors. 2001;14(1-4):69-74. doi: 10.1002/biof.5520140110. Biofactors. 2001. PMID: 11568442 Review.
Cited by
-
Biotransformation of Selenium by Lactic Acid Bacteria: Formation of Seleno-Nanoparticles and Seleno-Amino Acids.Front Bioeng Biotechnol. 2020 Jun 12;8:506. doi: 10.3389/fbioe.2020.00506. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32596220 Free PMC article.
-
Post-translational activation of non-selenium glutathione peroxidase of Chlamydomonas reinhardtii by specific incorporation of selenium.Biochem Biophys Rep. 2015 Aug 28;4:39-43. doi: 10.1016/j.bbrep.2015.08.018. eCollection 2015 Dec. Biochem Biophys Rep. 2015. PMID: 29124185 Free PMC article.
-
Increased Urinary 3-Mercaptolactate Excretion and Enhanced Passive Systemic Anaphylaxis in Mice Lacking Mercaptopyruvate Sulfurtransferase, a Model of Mercaptolactate-Cysteine Disulfiduria.Int J Mol Sci. 2020 Jan 27;21(3):818. doi: 10.3390/ijms21030818. Int J Mol Sci. 2020. PMID: 32012740 Free PMC article.
-
Selenium Nanoparticle-Enriched and Potential Probiotic, Lactiplantibacillus plantarum S14 Strain, a Diet Supplement Beneficial for Rainbow Trout.Biology (Basel). 2022 Oct 18;11(10):1523. doi: 10.3390/biology11101523. Biology (Basel). 2022. PMID: 36290428 Free PMC article.
-
Biochemical and biophysical characterization of the selenium-binding and reducing site in Arabidopsis thaliana homologue to mammals selenium-binding protein 1.J Biol Chem. 2014 Nov 14;289(46):31765-31776. doi: 10.1074/jbc.M114.571208. Epub 2014 Oct 1. J Biol Chem. 2014. PMID: 25274629 Free PMC article.
References
-
- Bock, A. & Stadtman, T. C. (1998) Biofactors 1, 245–250. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

