The protein structure prediction problem could be solved using the current PDB library
- PMID: 15653774
- PMCID: PMC545829
- DOI: 10.1073/pnas.0407152101
The protein structure prediction problem could be solved using the current PDB library
Abstract
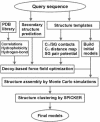
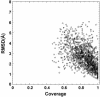
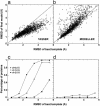
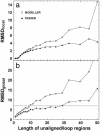
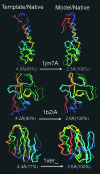
For single-domain proteins, we examine the completeness of the structures in the current Protein Data Bank (PDB) library for use in full-length model construction of unknown sequences. To address this issue, we employ a comprehensive benchmark set of 1,489 medium-size proteins that cover the PDB at the level of 35% sequence identity and identify templates by structure alignment. With homologous proteins excluded, we can always find similar folds to native with an average rms deviation (RMSD) from native of 2.5 A with approximately 82% alignment coverage. These template structures often contain a significant number of insertions/deletions. The tasser algorithm was applied to build full-length models, where continuous fragments are excised from the top-scoring templates and reassembled under the guide of an optimized force field, which includes consensus restraints taken from the templates and knowledge-based statistical potentials. For almost all targets (except for 2/1,489), the resultant full-length models have an RMSD to native below 6 A (97% of them below 4 A). On average, the RMSD of full-length models is 2.25 A, with aligned regions improved from 2.5 A to 1.88 A, comparable with the accuracy of low-resolution experimental structures. Furthermore, starting from state-of-the-art structural alignments, we demonstrate a methodology that can consistently bring template-based alignments closer to native. These results are highly suggestive that the protein-folding problem can in principle be solved based on the current PDB library by developing efficient fold recognition algorithms that can recover such initial alignments.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

