Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii
- PMID: 15653810
- PMCID: PMC1065355
- DOI: 10.1104/pp.104.055244
Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii
Abstract
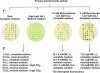
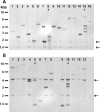
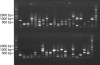
The unicellular green alga Chlamydomonas reinhardtii is a widely used model organism for studies of oxygenic photosynthesis in eukaryotes. Here we describe the development of a resource for functional genomics of photosynthesis using insertional mutagenesis of the Chlamydomonas nuclear genome. Chlamydomonas cells were transformed with either of two plasmids conferring zeocin resistance, and insertional mutants were selected in the dark on acetate-containing medium to recover light-sensitive and nonphotosynthetic mutants. The population of insertional mutants was subjected to a battery of primary and secondary phenotypic screens to identify photosynthesis-related mutants that were pigment deficient, light sensitive, nonphotosynthetic, or hypersensitive to reactive oxygen species. Approximately 9% of the insertional mutants exhibited 1 or more of these phenotypes. Molecular analysis showed that each mutant line contains an average of 1.4 insertions, and genetic analysis indicated that approximately 50% of the mutations are tagged by the transforming DNA. Flanking DNA was isolated from the mutants, and sequence data for the insertion sites in 50 mutants are presented and discussed.
Figures

References
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Asamizu E, Nakmura Y, Sato S, Fukuzawa H, Tabata S (1999) A large scale structural analysis of cDNAs in a unicellular green alga, Chlamydomonas reinhardtii. I. Generation of 3433 non-redundant expressed sequence tags. DNA Res 6: 369–373 - PubMed
-
- Barakat A, Gallois P, Raynal M, Mestre-Ortega D, Sallaud C, Guiderdoni E, Delseny M, Bernardi G (2000) The distribution of T-DNA in the genomes of transgenic Arabidopsis and rice. FEBS Lett 471: 161–164 - PubMed
-
- Chen SY, Jin WZ, Wang MY, Zhang F, Zhou J, Jia OJ, Wu YR, Liu FY, Wu P (2003) Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J 36: 105–113 - PubMed
-
- Clarke L, Carbon J (1976) A colony bank containing synthetic Col-E-1 hybrid plasmids representative of the entire Escherichia coli genome. Cell 9: 91–99 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

