Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens
- PMID: 15653821
- PMCID: PMC544178
- DOI: 10.1128/CMR.18.1.102-127.2005
Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens
Abstract
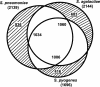
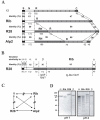
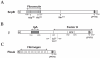
Streptococcus agalactiae (group B Streptococcus) is the major cause of invasive bacterial disease, including meningitis, in the neonatal period. Although prophylactic measures have contributed to a substantial reduction in the number of infections, development of a vaccine remains an important goal. While much work in this field has focused on the S. agalactiae polysaccharide capsule, which is an important virulence factor that elicits protective immunity, surface proteins have received increasing attention as potential virulence factors and vaccine components. Here, we summarize current knowledge about S. agalactiae surface proteins, with emphasis on proteins that have been characterized immunochemically and/or elicit protective immunity in animal models. These surface proteins have been implicated in interactions with human epithelial cells, binding to extracellular matrix components, and/or evasion of host immunity. Of note, several S. agalactiae surface proteins are related to surface proteins identified in other bacterial pathogens, emphasizing the general interest of the S. agalactiae proteins. Because some S. agalactiae surface proteins elicit protective immunity, they hold promise as components in a vaccine based only on proteins or as carriers in polysaccharide conjugate vaccines.
Figures


Similar articles
-
Identification and immunoreactivity of proteins released from Streptococcus agalactiae.Eur J Clin Microbiol Infect Dis. 2004 Nov;23(11):818-24. doi: 10.1007/s10096-004-1229-y. Epub 2004 Oct 14. Eur J Clin Microbiol Infect Dis. 2004. PMID: 15490293
-
Glutathione Synthesis Contributes to Virulence of Streptococcus agalactiae in a Murine Model of Sepsis.J Bacteriol. 2019 Sep 20;201(20):e00367-19. doi: 10.1128/JB.00367-19. Print 2019 Oct 15. J Bacteriol. 2019. PMID: 31331978 Free PMC article.
-
Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease.Mol Microbiol. 2002 Sep;45(6):1499-513. doi: 10.1046/j.1365-2958.2002.03126.x. Mol Microbiol. 2002. PMID: 12354221
-
Bacterial virulence factors in neonatal sepsis: group B streptococcus.Curr Opin Infect Dis. 2004 Jun;17(3):225-9. doi: 10.1097/00001432-200406000-00009. Curr Opin Infect Dis. 2004. PMID: 15166825 Review.
-
Streptococcus agalactiae Non-Pilus, Cell Wall-Anchored Proteins: Involvement in Colonization and Pathogenesis and Potential as Vaccine Candidates.Front Immunol. 2018 Apr 5;9:602. doi: 10.3389/fimmu.2018.00602. eCollection 2018. Front Immunol. 2018. PMID: 29686667 Free PMC article. Review.
Cited by
-
Listeria monocytogenes surface proteins: from genome predictions to function.Microbiol Mol Biol Rev. 2007 Jun;71(2):377-97. doi: 10.1128/MMBR.00039-06. Microbiol Mol Biol Rev. 2007. PMID: 17554049 Free PMC article. Review.
-
Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition.Genome Biol. 2007;8(5):R71. doi: 10.1186/gb-2007-8-5-r71. Genome Biol. 2007. PMID: 17475002 Free PMC article.
-
Host and pathogen glycosaminoglycan-binding proteins modulate antimicrobial peptide responses in Drosophila melanogaster.Infect Immun. 2011 Feb;79(2):606-16. doi: 10.1128/IAI.00254-10. Epub 2010 Nov 15. Infect Immun. 2011. PMID: 21078848 Free PMC article.
-
Emergence of respiratory Streptococcus agalactiae isolates in cystic fibrosis patients.PLoS One. 2009;4(2):e4650. doi: 10.1371/journal.pone.0004650. Epub 2009 Feb 27. PLoS One. 2009. PMID: 19247473 Free PMC article.
-
Group B Streptococcal Colonization, Molecular Characteristics, and Epidemiology.Front Microbiol. 2018 Mar 14;9:437. doi: 10.3389/fmicb.2018.00437. eCollection 2018. Front Microbiol. 2018. PMID: 29593684 Free PMC article. Review.
References
-
- Anderson, E. T., M. G. Wetherell, L. A. Winter, S. B. Olmsted, P. P. Cleary, and Y. V. Matsuka. 2002. Processing, stability, and kinetic parameters of C5a peptidase from Streptococcus pyogenes. Eur. J. Biochem. 269:4839-4851. - PubMed
-
- Areschoug, T., F. Carlsson, M. Stålhammar-Carlemalm, and G. Lindahl. 2004. Host-pathogen interactions in Streptococcus pyogenes infections, with special reference to puerperal fever and a comment on vaccine development. Vaccine 22:S9-S14. - PubMed
-
- Areschoug, T., M. Stålhammar-Carlemalm, I. Karlsson, and G. Lindahl. 2002. Streptococcal β protein has separate binding sites for human factor H and IgA-Fc. J. Biol. Chem. 277:12642-12648. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

