Divergent V1R repertoires in five species: Amplification in rodents, decimation in primates, and a surprisingly small repertoire in dogs
- PMID: 15653832
- PMCID: PMC546524
- DOI: 10.1101/gr.3339905
Divergent V1R repertoires in five species: Amplification in rodents, decimation in primates, and a surprisingly small repertoire in dogs
Erratum in
- Genome Res. 2005 Apr;15(4):601
Abstract
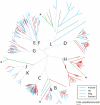
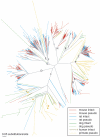
The V1R gene family comprises one of two types of putative pheromone receptors expressed in the mammalian vomeronasal organ (VNO). We searched the most recent mouse, rat, dog, chimpanzee, and human genome sequence assemblies to compile a near-complete repertoire of V1R genes for each species. Dog, human, and chimpanzee have very few intact V1Rs (8, 2, and 0, respectively) compared to more than a hundred intact V1Rs in each of the rat (106) and mouse (165) genomes. We also provide the first description of the diversity of V1R pseudogenes in these species. We identify at least 165 pseudogenes in mouse, 110 in rat, 102 in chimpanzee, 115 in human, and 54 in dog. Primate and dog pseudogenes are distributed among almost all V1R subfamilies seen in rodents, indicating that the common ancestor of these species had a diverse V1R repertoire. We find that V1R genes were subject to strikingly different fates in different species and in different subfamilies. In rodents, some subfamilies remained relatively stable or underwent roughly equivalent expansion in mouse and rat; other subfamilies expanded in one species but not the other. The small number of intact V1Rs in the dog genome is unexpected given the presumption that dogs, like rodents, have a functional VNO, and a complex system of pheromone-based behaviors. We identify an intact transient receptor potential channel 2beta in the dog genome, consistent with a functional VNO in dogs. The diminished V1R repertoire in dogs raises questions about the relative contributions of V1Rs versus other candidate pheromone receptor genes in the establishment of complex pheromone systems in mammals.
Figures
References
-
- Boschat, C., Pelofi, C., Randin, O., Roppolo, D., Luscher, C., Broillet, M.-C., and Rodriguez, I. 2002. Pheromone detection mediated by a V1r vomeronasal receptor. Nat. Neurosci. 5: 1261-1262. - PubMed
-
- Buck, L. and Axel, R. 1991. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 65: 175-187. - PubMed
-
- Cohen-Tannoudji, J., Lavenet, C., Locatelli, A., Tillet, Y., and Signoret, J.P. 1989. Non-involvement of the accessory olfactory system in the LH response of anoestrous ewes to male odour. J. Reprod. Fertil. 86: 135-144. - PubMed
-
- Dehal, P., Predki, P., Olsen, A.S., Kobayashi, A., Folta, P., Lucas, S., Land, M., Terry, A., Ecale Zhou, C.L., Rash, S., et al. 2001. Human chromosome 19 and related regions in mouse: Conservative and lineage-specific evolution. Science 293: 104-111. - PubMed
-
- Del Punta, K., Leinders-Zufall, T., Rodriguez, I., Jukam, D., Wysocki, C.J., Ogawa, S., Zufall, F., and Mombaerts, P. 2002. Deficient pheromone responses in mice lacking a cluster of vomeronasal receptor genes. Nature 419: 70-74. - PubMed
WEB SITE REFERENCES
-
- http://ftp.genome.washington.edu/cgi-bin/RepeatMasker; RepeatMasker Web server.
-
- http://genome.ucsc.edu/; UCSC Genome Bioinformatics.
-
- http://hiv-web.lanl.gov/content/hiv-db/SNAP/WEBSNAP/SNAP.html; HIV Sequence Database: SNAP submission form.
-
- http://repeatmasker.org; RepeatMasker.
-
- http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html; BCM search launcher.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
