The heme activator protein Hap1 represses transcription by a heme-independent mechanism in Saccharomyces cerevisiae
- PMID: 15654089
- PMCID: PMC1449556
- DOI: 10.1534/genetics.104.037143
The heme activator protein Hap1 represses transcription by a heme-independent mechanism in Saccharomyces cerevisiae
Abstract
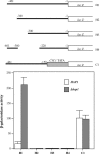
The yeast heme activator protein Hap1 binds to DNA and activates transcription of genes encoding functions required for respiration and for controlling oxidative damage, in response to heme. Hap1 contains a DNA-binding domain with a C6 zinc cluster motif, a coiled-coil dimerization element, typical of the members of the yeast Gal4 family, and an acidic activation domain. The regulation of Hap1 transcription-activating activity is controlled by two classes of Hap1 elements, repression modules (RPM1-3) and heme-responsive motifs (HRM1-7). Previous indirect evidence indicates that Hap1 may repress transcription directly. Here we show, by promoter analysis, by chromatin immunoprecipitation, and by electrophoretic mobility shift assay, that Hap1 binds directly to DNA and represses transcription of its own gene by at least 20-fold. We found that Hap1 repression of the HAP1 gene occurs independently of heme concentrations. While DNA binding is required for transcriptional repression by Hap1, deletion of Hap1 activation domain and heme-regulatory elements has varying effects on repression. Further, we found that repression by Hap1 requires the function of Hsp70 (Ssa), but not Hsp90. These results show that Hap1 binds to its own promoter and represses transcription in a heme-independent but Hsp70-dependent manner.
Figures



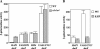
Similar articles
-
The Hsp70-Ydj1 molecular chaperone represses the activity of the heme activator protein Hap1 in the absence of heme.Mol Cell Biol. 2001 Dec;21(23):7923-32. doi: 10.1128/MCB.21.23.7923-7932.2001. Mol Cell Biol. 2001. PMID: 11689685 Free PMC article.
-
Heme levels switch the function of Hap1 of Saccharomyces cerevisiae between transcriptional activator and transcriptional repressor.Mol Cell Biol. 2007 Nov;27(21):7414-24. doi: 10.1128/MCB.00887-07. Epub 2007 Sep 4. Mol Cell Biol. 2007. PMID: 17785431 Free PMC article.
-
Functional analysis of heme regulatory elements of the transcriptional activator Hap1.Biochem Biophys Res Commun. 2000 Jul 5;273(2):584-91. doi: 10.1006/bbrc.2000.2995. Biochem Biophys Res Commun. 2000. PMID: 10873649
-
A unique mechanism of chaperone action: heme regulation of Hap1 activity involves separate control of repression and activation.Protein Pept Lett. 2009;16(6):642-9. doi: 10.2174/092986609788490113. Protein Pept Lett. 2009. PMID: 19519523 Review.
-
Molecular mechanism of heme signaling in yeast: the transcriptional activator Hap1 serves as the key mediator.Cell Mol Life Sci. 1999 Oct 30;56(5-6):415-26. doi: 10.1007/s000180050442. Cell Mol Life Sci. 1999. PMID: 11212295 Free PMC article. Review.
Cited by
-
Antifungal activity of berberine hydrochloride and palmatine hydrochloride against Microsporum canis -induced dermatitis in rabbits and underlying mechanism.BMC Complement Altern Med. 2015 Jun 9;15:177. doi: 10.1186/s12906-015-0680-x. BMC Complement Altern Med. 2015. PMID: 26054937 Free PMC article.
-
A fungal family of transcriptional regulators: the zinc cluster proteins.Microbiol Mol Biol Rev. 2006 Sep;70(3):583-604. doi: 10.1128/MMBR.00015-06. Microbiol Mol Biol Rev. 2006. PMID: 16959962 Free PMC article. Review.
-
Heme promotes transcriptional and demethylase activities of Gis1, a member of the histone demethylase JMJD2/KDM4 family.Nucleic Acids Res. 2018 Jan 9;46(1):215-228. doi: 10.1093/nar/gkx1051. Nucleic Acids Res. 2018. PMID: 29126261 Free PMC article.
-
Impact of DNA-binding position variants on yeast gene expression.Nucleic Acids Res. 2009 Nov;37(21):6991-7001. doi: 10.1093/nar/gkp743. Nucleic Acids Res. 2009. PMID: 19767613 Free PMC article.
-
Further support for aneuploidy tolerance in wild yeast and effects of dosage compensation on gene copy-number evolution.Elife. 2016 Mar 7;5:e14409. doi: 10.7554/eLife.14409. Elife. 2016. PMID: 26949252 Free PMC article.
References
-
- Buisson, N., and R. Labbe-Bois, 1998. Flavohemoglobin expression and function in Saccharomyces cerevisiae. No relationship with respiration and complex response to oxidative stress. J. Biol. Chem. 273: 9527–9533. - PubMed
-
- Chang, H. C., and S. Lindquist, 1994. Conservation of Hsp90 macromolecular complexes in Saccharomyces cerevisiae. J. Biol. Chem. 269: 24983–24988. - PubMed
-
- Choi, J. Y., J. Stukey, S. Y. Hwang and C. E. Martin, 1996. Regulatory elements that control transcription activation and unsaturated fatty acid-mediated repression of the Saccharomyces cerevisiae OLE1 gene. J. Biol. Chem. 271: 3581–3589. - PubMed
-
- Creusot, F., J. Verdiere, M. Gaisne and P. P. Slonimski, 1988. CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. I. Overall organization of the protein sequence displays several novel structural domains. J. Mol. Biol. 204: 263–276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

