Meiosis-specific regulation of the Saccharomyces cerevisiae S-phase cyclin CLB5 is dependent on MluI cell cycle box (MCB) elements in its promoter but is independent of MCB-binding factor activity
- PMID: 15654101
- PMCID: PMC1449548
- DOI: 10.1534/genetics.104.036103
Meiosis-specific regulation of the Saccharomyces cerevisiae S-phase cyclin CLB5 is dependent on MluI cell cycle box (MCB) elements in its promoter but is independent of MCB-binding factor activity
Abstract
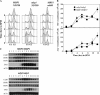
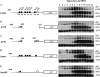
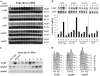
In proliferating S. cerevisiae, genes whose products function in DNA replication are regulated by the MBF transcription factor composed of Mbp1 and Swi6 that binds to consensus MCB sequences in target promoters. We find that during meiotic development a subset of DNA replication genes exemplified by TMP1 and RNR1 are regulated by Mbp1. Deletion of Mbp1 deregulated TMP1 and RNR1 but did not interfere with premeiotic S-phase, meiotic recombination, or spore formation. Surprisingly, deletion of MBP1 had no effect on the expression of CLB5, which is purportedly controlled by MBF. Extensive analysis of the CLB5 promoter revealed that the gene is largely regulated by elements within a 100-bp fragment containing a cluster of MCB sequences. Surprisingly, induction of the CLB5 promoter requires MCB sequences, but not Mbp1, implying that another MCB-binding factor may exist in cells undergoing meiosis. In addition, full activation of CLB5 during meiosis requires Clb5 activity, suggesting that CLB5 may be regulated by a positive feedback mechanism. We further demonstrate that during meiosis MCBs function as effective transcriptional activators independent of MBP1.
Figures



Similar articles
-
The Saccharomyces cerevisiae CLB5 promoter contains two middle sporulation elements (MSEs) that are differentially regulated during sporulation.Yeast. 2008 Apr;25(4):259-72. doi: 10.1002/yea.1585. Yeast. 2008. PMID: 18327887
-
Set1- and Clb5-deficiencies disclose the differential regulation of centromere and telomere dynamics in Saccharomyces cerevisiae meiosis.J Cell Sci. 2005 Nov 1;118(Pt 21):4985-94. doi: 10.1242/jcs.02612. J Cell Sci. 2005. PMID: 16254243
-
A central role for SWI6 in modulating cell cycle Start-specific transcription in yeast.Nature. 1992 Jun 11;357(6378):508-13. doi: 10.1038/357508a0. Nature. 1992. PMID: 1608451
-
DNA synthesis control in yeast: an evolutionarily conserved mechanism for regulating DNA synthesis genes?Bioessays. 1992 Dec;14(12):823-30. doi: 10.1002/bies.950141206. Bioessays. 1992. PMID: 1365898 Review.
-
Cell cycle control and initiation of DNA replication in Saccharomyces cerevisiae.Biol Chem. 1996 Jul-Aug;377(7-8):481-7. Biol Chem. 1996. PMID: 8922282 Review.
Cited by
-
Ume6 Acts as a Stable Platform To Coordinate Repression and Activation of Early Meiosis-Specific Genes in Saccharomyces cerevisiae.Mol Cell Biol. 2021 Jun 23;41(7):e0037820. doi: 10.1128/MCB.00378-20. Epub 2021 Jun 23. Mol Cell Biol. 2021. PMID: 33941619 Free PMC article.
-
A switch from a gradient to a threshold mode in the regulation of a transcriptional cascade promotes robust execution of meiosis in budding yeast.PLoS One. 2010 Jun 8;5(6):e11005. doi: 10.1371/journal.pone.0011005. PLoS One. 2010. PMID: 20543984 Free PMC article.
-
Polysaccharides derived from Balanophora polyandra significantly suppressed the proliferation of ovarian cancer cells through P53-mediated pathway.J Cell Mol Med. 2020 Jul;24(14):8115-8125. doi: 10.1111/jcmm.15468. Epub 2020 Jun 10. J Cell Mol Med. 2020. PMID: 32519803 Free PMC article.
-
Execution of the meiotic noncoding RNA expression program and the onset of gametogenesis in yeast require the conserved exosome subunit Rrp6.Proc Natl Acad Sci U S A. 2011 Jan 18;108(3):1058-63. doi: 10.1073/pnas.1016459108. Epub 2010 Dec 13. Proc Natl Acad Sci U S A. 2011. PMID: 21149693 Free PMC article.
-
Phosphorylation and dephosphorylation regulate APC/C(Cdh1) substrate degradation.Cell Cycle. 2015;14(19):3138-45. doi: 10.1080/15384101.2015.1078036. Cell Cycle. 2015. PMID: 26252546 Free PMC article.
References
-
- Borde, V., A. S. H. Goldman and M. Lichten, 2000. Direct coupling between meiotic DNA replication and recombination initiation. Science 290: 806–809. - PubMed
-
- Breeden, L., and K. Nasmyth, 1987. Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell 48: 389–397. - PubMed
-
- Cho, R. J., M. J. Campbell, E. A. Winzeler, L. Steinmetz, A. Conway et al., 1998. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 2: 65–73. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

