Statistical tests of the coalescent model based on the haplotype frequency distribution and the number of segregating sites
- PMID: 15654103
- PMCID: PMC1449527
- DOI: 10.1534/genetics.104.032219
Statistical tests of the coalescent model based on the haplotype frequency distribution and the number of segregating sites
Abstract
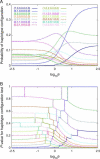
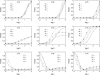
Several tests of neutral evolution employ the observed number of segregating sites and properties of the haplotype frequency distribution as summary statistics and use simulations to obtain rejection probabilities. Here we develop a "haplotype configuration test" of neutrality (HCT) based on the full haplotype frequency distribution. To enable exact computation of rejection probabilities for small samples, we derive a recursion under the standard coalescent model for the joint distribution of the haplotype frequencies and the number of segregating sites. For larger samples, we consider simulation-based approaches. The utility of the HCT is demonstrated in simulations of alternative models and in application to data from Drosophila melanogaster.
Figures

References
-
- Abramowitz, M. A., and I. A. Stegun, 1965 Handbook of Mathematical Functions. Dover, New York.
-
- Begun, D. J., and C. F. Aquadro, 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in Drosophila melanogaster. Nature 356: 519–520. - PubMed
-
- Berger, R. L., and D. D. Boos, 1994. P values maximized over a confidence set for the nuisance parameter. J. Am. Stat. Assoc. 89: 1012–1016.
-
- Depaulis, F., and M. Veuille, 1998. Neutrality tests based on the distribution of haplotypes under an infinite-site model. Mol. Biol. Evol. 15: 1788–1790. - PubMed
-
- Depaulis, F., S. Mousset and M. Veuille, 2001. Haplotype tests using coalescent simulations conditional on the number of segregating sites. Mol. Biol. Evol. 18: 1136–1138. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

