Tissue-specific, nutritional, and developmental regulation of rat fatty acid elongases
- PMID: 15654130
- PMCID: PMC2430181
- DOI: 10.1194/jlr.M400335-JLR200
Tissue-specific, nutritional, and developmental regulation of rat fatty acid elongases
Abstract
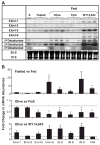
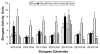
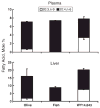
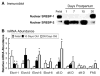
Of the six fatty acid elongase (Elovl) subtypes expressed in mammals, adult rat liver expresses four subtypes: Elovl-5 > Elovl-1 = Elovl-2 = Elovl-6. Overnight starvation and fish oil-enriched diets repressed hepatic elongase activity in livers of adult male rats. Diet-induced changes in elongase activity correlate with Elovl-5 and Elovl-6 mRNA abundance. Adult rats fed the peroxisome proliferator-activated receptor alpha (PPARalpha) agonist WY14,643 have increased hepatic elongase activity, Elovl-1, Elovl-5, Elovl-6, Delta5, Delta6, and Delta9 desaturase mRNA abundance, and mead acid (20:3,n-9) content. PPARalpha agonists affect both fatty acid elongation and desaturation pathways leading to changes in hepatic lipid composition. Elovl activity is low in fetal liver but increases significantly after birth. Developmental changes in hepatic elongase activity paralleled the postnatal induction of Elovl-5 mRNA and mRNAs encoding the PPARalpha-regulated transcripts, Delta5 and Delta6 desaturase, and cytochrome P450 4A. In contrast, Elovl-6, Delta9 desaturase, and FAS mRNA abundance paralleled changes in hepatic sterol regulatory element binding protein 1c (SREBP-1c) nuclear content. SREBP-1c is present in fetal liver nuclei, absent from nuclei immediately after birth, and reappears in nuclei at weaning, 21 days postpartum. In conclusion, changes in Elovl-5 expression may account for much of the nutritional and developmental control of fatty acid elongation activity in the rat liver.
Figures


Similar articles
-
Regulation of hepatic fatty acid elongase and desaturase expression in diabetes and obesity.J Lipid Res. 2006 Sep;47(9):2028-41. doi: 10.1194/jlr.M600177-JLR200. Epub 2006 Jun 21. J Lipid Res. 2006. PMID: 16790840 Free PMC article.
-
Role of fatty acid elongases in determination of de novo synthesized monounsaturated fatty acid species.J Lipid Res. 2010 Jul;51(7):1871-7. doi: 10.1194/jlr.M004747. Epub 2010 Mar 12. J Lipid Res. 2010. PMID: 20228221 Free PMC article.
-
Elevated hepatic fatty acid elongase-5 activity affects multiple pathways controlling hepatic lipid and carbohydrate composition.J Lipid Res. 2008 Jul;49(7):1538-52. doi: 10.1194/jlr.M800123-JLR200. Epub 2008 Mar 30. J Lipid Res. 2008. PMID: 18376007 Free PMC article.
-
Fatty acid regulation of hepatic gene transcription.J Nutr. 2005 Nov;135(11):2503-6. doi: 10.1093/jn/135.11.2503. J Nutr. 2005. PMID: 16251601 Review.
-
The ELOVL proteins: Very and ultra long-chain fatty acids at the crossroads between metabolic and neurodegenerative disorders.Mol Genet Metab. 2025 Mar;144(3):109050. doi: 10.1016/j.ymgme.2025.109050. Epub 2025 Feb 4. Mol Genet Metab. 2025. PMID: 39946831 Review.
Cited by
-
Examining the role of lipid mediators in diabetic retinopathy.Clin Lipidol. 2012 Dec 1;7(6):661-675. doi: 10.2217/clp.12.68. Clin Lipidol. 2012. PMID: 23646066 Free PMC article.
-
The gut microbiota promotes hepatic fatty acid desaturation and elongation in mice.Nat Commun. 2018 Sep 14;9(1):3760. doi: 10.1038/s41467-018-05767-4. Nat Commun. 2018. PMID: 30218046 Free PMC article.
-
The influence of maternal early to mid-gestation nutrient restriction on long chain polyunsaturated fatty acids in fetal sheep.Lipids. 2008 Jun;43(6):525-31. doi: 10.1007/s11745-008-3186-1. Epub 2008 May 15. Lipids. 2008. PMID: 18481131
-
Diet Regulation of Long-Chain PUFA Synthesis: Role of Macronutrients, Micronutrients, and Polyphenols on Δ-5/Δ-6 Desaturases and Elongases 2/5.Adv Nutr. 2021 Jun 1;12(3):980-994. doi: 10.1093/advances/nmaa142. Adv Nutr. 2021. PMID: 33186986 Free PMC article. Review.
-
Hepatic and plasma sex differences in saturated and monounsaturated fatty acids are associated with differences in expression of elongase 6, but not stearoyl-CoA desaturase in Sprague-Dawley rats.Genes Nutr. 2013 May;8(3):317-27. doi: 10.1007/s12263-012-0325-3. Epub 2012 Nov 22. Genes Nutr. 2013. PMID: 23180365 Free PMC article.
References
-
- Jump DB, Clarke SD. Regulation of gene expression by dietary fat. Annu Rev Nutr. 1999;19:63–90. - PubMed
-
- Jump D. Fatty acid regulation of gene transcription. Crit Rev Clin Lab Sci. 2004;41:41–78. - PubMed
-
- Hillgartner FB, Salati LM, Goodridge AG. Physiological and molecular mechanisms involved in nutritional regulation of fatty acid synthesis. Physiol Rev. 1995;75:47–76. - PubMed
-
- Pawar A, Xu J, Jerks E, Mangelsdorf DJ, Jump DB. Fatty acid regulation of liver X receptors (LXR) and peroxisome proliferator-activated receptor alpha (PPARalpha) in HEK293 cells. J Biol Chem. 2002;277:39243–39250. - PubMed
-
- Pawar A, Jump DB. Unsaturated fatty acid regulation of PPAR-alpha in rat primary hepatocytes. J Biol Chem. 2003;278:35931–35939. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

