Pi class glutathione S-transferase genes are regulated by Nrf 2 through an evolutionarily conserved regulatory element in zebrafish
- PMID: 15654768
- PMCID: PMC1186694
- DOI: 10.1042/BJ20041860
Pi class glutathione S-transferase genes are regulated by Nrf 2 through an evolutionarily conserved regulatory element in zebrafish
Abstract
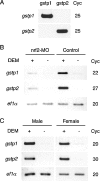
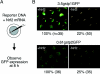
Pi class GSTs (glutathione S-transferases) are a member of the vertebrate GST family of proteins that catalyse the conjugation of GSH to electrophilic compounds. The expression of Pi class GST genes can be induced by exposure to electrophiles. We demonstrated previously that the transcription factor Nrf 2 (NF-E2 p45-related factor 2) mediates this induction, not only in mammals, but also in fish. In the present study, we have isolated the genomic region of zebrafish containing the genes gstp1 and gstp2. The regulatory regions of zebrafish gstp1 and gstp2 have been examined by GFP (green fluorescent protein)-reporter gene analyses using microinjection into zebrafish embryos. Deletion and point-mutation analyses of the gstp1 promoter showed that an ARE (antioxidant-responsive element)-like sequence is located 50 bp upstream of the transcription initiation site which is essential for Nrf 2 transactivation. Using EMSA (electrophoretic mobility-shift assay) analysis we showed that zebrafish Nrf 2-MafK heterodimer specifically bound to this sequence. All the vertebrate Pi class GST genes harbour a similar ARE-like sequence in their promoter regions. We propose that this sequence is a conserved target site for Nrf 2 in the Pi class GST genes.
Figures





Similar articles
-
Activation of mouse Pi-class glutathione S-transferase gene by Nrf2(NF-E2-related factor 2) and androgen.Biochem J. 2002 Jun 1;364(Pt 2):563-70. doi: 10.1042/BJ20011756. Biochem J. 2002. PMID: 12023900 Free PMC article.
-
Molecular identification of glutathione S-transferase gene and cDNAs of two isotypes from northern quahog (Mercenaria mercenaria).Comp Biochem Physiol B Biochem Mol Biol. 2009 Sep;154(1):25-36. doi: 10.1016/j.cbpb.2009.04.012. Epub 2009 May 3. Comp Biochem Physiol B Biochem Mol Biol. 2009. PMID: 19410653
-
The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance.Crit Rev Biochem Mol Biol. 1995;30(6):445-600. doi: 10.3109/10409239509083491. Crit Rev Biochem Mol Biol. 1995. PMID: 8770536 Review.
-
Characterization of glutathione-S-transferases in zebrafish (Danio rerio).Aquat Toxicol. 2015 Jan;158:50-62. doi: 10.1016/j.aquatox.2014.10.013. Epub 2014 Nov 6. Aquat Toxicol. 2015. PMID: 25461745
-
Mammalian cytosolic glutathione transferases.Curr Protein Pept Sci. 2008 Aug;9(4):325-37. doi: 10.2174/138920308785132677. Curr Protein Pept Sci. 2008. PMID: 18691123 Review.
Cited by
-
Embryo Microinjection of Selenomethionine Reduces Hatchability and Modifies Oxidant Responsive Gene Expression in Zebrafish.Sci Rep. 2016 May 23;6:26520. doi: 10.1038/srep26520. Sci Rep. 2016. PMID: 27210033 Free PMC article.
-
Marine glutathione S-transferases.Mar Biotechnol (NY). 2007 Sep-Oct;9(5):513-42. doi: 10.1007/s10126-007-9034-0. Epub 2007 Aug 9. Mar Biotechnol (NY). 2007. PMID: 17682821 Review.
-
Nrf2 activation attenuates genetic endoplasmic reticulum stress induced by a mutation in the phosphomannomutase 2 gene in zebrafish.Proc Natl Acad Sci U S A. 2018 Mar 13;115(11):2758-2763. doi: 10.1073/pnas.1714056115. Epub 2018 Feb 22. Proc Natl Acad Sci U S A. 2018. PMID: 29472449 Free PMC article.
-
Nrf2b, novel zebrafish paralog of oxidant-responsive transcription factor NF-E2-related factor 2 (NRF2).J Biol Chem. 2012 Feb 10;287(7):4609-27. doi: 10.1074/jbc.M111.260125. Epub 2011 Dec 15. J Biol Chem. 2012. PMID: 22174413 Free PMC article.
-
Transcriptome analysis reveals the importance of exogenous nutrition in regulating antioxidant defenses during the mouth-opening stage in oviparous fish.Fish Physiol Biochem. 2021 Aug;47(4):1087-1103. doi: 10.1007/s10695-021-00954-5. Epub 2021 May 25. Fish Physiol Biochem. 2021. PMID: 34036482
References
-
- Itoh K., Tong K. I., Yamamoto M. Molecular mechanism activating Nrf 2–Keap1 pathway in regulation of adaptive response to electrophiles. Free Radical Biol. Med. 2004;36:1208–1213. - PubMed
-
- Kobayashi M., Yamamoto M. Molecular mechanisms activating the Nrf 2–Keap1 pathway of antioxidant gene regulation. Antioxid. Redox Signal. 2005;7:385–394. - PubMed
-
- Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., et al. An Nrf 2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

