Virus factories: associations of cell organelles for viral replication and morphogenesis
- PMID: 15656780
- PMCID: PMC7161905
- DOI: 10.1042/BC20040058
Virus factories: associations of cell organelles for viral replication and morphogenesis
Abstract
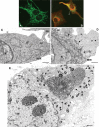
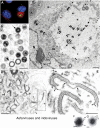
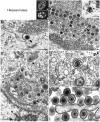
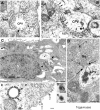
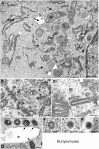
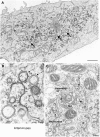
Genome replication and assembly of viruses often takes place in specific intracellular compartments where viral components concentrate, thereby increasing the efficiency of the processes. For a number of viruses the formation of 'factories' has been described, which consist of perinuclear or cytoplasmic foci that mostly exclude host proteins and organelles but recruit specific cell organelles, building a unique structure. The formation of the viral factory involves a number of complex interactions and signalling events between viral and cell factors. Mitochondria, cytoplasmic membranes and cytoskeletal components frequently participate in the formation of viral factories, supplying basic and common needs for key steps in the viral replication cycle.
Figures


References
-
- Baxter J. Merkenschlager M. Fisher A.G. Nuclear organisation and gene expression Curr. Opin. Cell Biol. 2002. 14 372–376. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

