Evidence for preferential copackaging of Moloney murine leukemia virus genomic RNAs transcribed in the same chromosomal site
- PMID: 15656910
- PMCID: PMC546228
- DOI: 10.1186/1742-4690-2-3
Evidence for preferential copackaging of Moloney murine leukemia virus genomic RNAs transcribed in the same chromosomal site
Abstract
Background: Retroviruses have a diploid genome and recombine at high frequency. Recombinant proviruses can be generated when two genetically different RNA genomes are packaged into the same retroviral particle. It was shown in several studies that recombinant proviruses could be generated in each round of HIV-1 replication, whereas the recombination rates of SNV and Mo-MuLV are 5 to 10-fold lower. The reason for these differences is not clear. One possibility is that these retroviruses may differ in their ability to copackage genomic RNAs produced at different chromosomal loci.
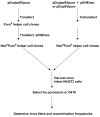
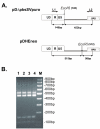
Results: To investigate whether there is a difference in the efficiency of heterodimer formation when two proviruses have the same or different chromosomal locations, we introduced two different Mo-MuLV-based retroviral vectors into the packaging cell line using either the cotransfection or sequential transfection procedure. The comparative study has shown that the frequency of recombination increased about four-fold when the cotransfection procedure was used. This difference was not associated with possible recombination of retroviral vectors during or after cotransfection and the ratios of retroviral virion RNAs were the same for two variants of transfection.
Conclusions: The results of this study indicate that a mechanism exists to enable the preferential copackaging of Mo-MuLV genomic RNA molecules that are transcribed on the same DNA template. The properties of Mo-MuLV genomic RNAs transport, processing or dimerization might be responsible for this preference. The data presented in this report can be useful when designing methods to study different aspects of replication and recombination of a diploid retroviral genome.
Figures


Similar articles
-
Homologous recombination of copackaged retrovirus RNAs during reverse transcription.J Virol. 1992 Apr;66(4):2378-88. doi: 10.1128/JVI.66.4.2378-2388.1992. J Virol. 1992. PMID: 1372369 Free PMC article.
-
Mutational analysis of stem-loops in the RNA packaging signal of the Moloney murine leukemia virus.Virology. 1998 Apr 25;244(1):133-45. doi: 10.1006/viro.1998.9090. Virology. 1998. PMID: 9581786
-
Complementarity-directed RNA dimer-linkage promotes retroviral recombination in vivo.Nucleic Acids Res. 2004 Jan 9;32(1):102-14. doi: 10.1093/nar/gkh159. Print 2004. Nucleic Acids Res. 2004. PMID: 14715920 Free PMC article.
-
How retroviruses select their genomes.Nat Rev Microbiol. 2005 Aug;3(8):643-55. doi: 10.1038/nrmicro1210. Nat Rev Microbiol. 2005. PMID: 16064056 Review.
-
Genetic reassortment and patch repair by recombination in retroviruses.J Biomed Sci. 2000 Mar-Apr;7(2):77-99. doi: 10.1007/BF02256615. J Biomed Sci. 2000. PMID: 10754383 Review.
Cited by
-
Suboptimal provirus expression explains apparent nonrandom cell coinfection with HIV-1.J Virol. 2012 Aug;86(16):8810-20. doi: 10.1128/JVI.00831-12. Epub 2012 Jun 13. J Virol. 2012. PMID: 22696639 Free PMC article.
-
Murine leukemia virus RNA dimerization is coupled to transcription and splicing processes.Retrovirology. 2010 Aug 5;7:64. doi: 10.1186/1742-4690-7-64. Retrovirology. 2010. PMID: 20687923 Free PMC article.
-
Nuclear trafficking of retroviral RNAs and Gag proteins during late steps of replication.Viruses. 2013 Nov 18;5(11):2767-95. doi: 10.3390/v5112767. Viruses. 2013. PMID: 24253283 Free PMC article. Review.
-
Packaging of host mY RNAs by murine leukemia virus may occur early in Y RNA biogenesis.J Virol. 2009 Dec;83(23):12526-34. doi: 10.1128/JVI.01219-09. Epub 2009 Sep 23. J Virol. 2009. PMID: 19776129 Free PMC article.
-
Mechanisms of human immunodeficiency virus type 2 RNA packaging: efficient trans packaging and selection of RNA copackaging partners.J Virol. 2011 Aug;85(15):7603-12. doi: 10.1128/JVI.00562-11. Epub 2011 May 25. J Virol. 2011. PMID: 21613401 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

