Hinderin, a five-domains protein including coiled-coil motifs that binds to SMC3
- PMID: 15656913
- PMCID: PMC547899
- DOI: 10.1186/1471-2121-6-3
Hinderin, a five-domains protein including coiled-coil motifs that binds to SMC3
Abstract
Background: The structural maintenance of chromosome proteins SMC1 and SMC3 play an important role in the maintenance of chromosomal integrity by preventing the premature separation of the sister chromatids at the onset of anaphase. The two proteins are constitutive components of the multimeric complex cohesin and form dimers by interacting at their central globular regions.
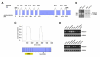
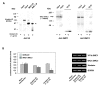
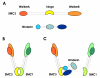
Results: In order to identify proteins that by binding to SMC3 may interfere with the protein dimerization process, a human cDNA library was screened by the yeast two-hybrid system by using the hinge region of SMC3 as bait. This has lead to the identification of Hinderin, a novel five domains protein including two coiled-coil motifs and sharing a strikingly structural similarity to the SMC family of proteins. Hinderin is ubiquitously expressed in human tissues. Orthologue forms of the protein are present in other vertebrates but not in lower organisms. A mapping of the interaction sites revealed that the N- and C-terminal globular domains mediate the binding of Hinderin to SMC3. Hinderin/SMC3 complexes could be recovered by immunoprecipitation from cell lysates using an anti-SMC3 antibody, thus demonstrating that the two proteins interact in vivo. On the contrary, Hinderin did not interact with SMC1. In vivo the rate of SMC1/SMC3 interaction was decreased by the ectopic expression of Hinderin.
Conclusions: Hinderin is a novel binding partner of SMC3. Based on its ability to modulate SMC1/SMC3 interaction we postulate that Hinderin affects the availability of SMC3 to engage in the formation of multimeric protein complexes.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

