Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor
- PMID: 15657141
- PMCID: PMC545828
- DOI: 10.1073/pnas.0404952102
Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor
Abstract
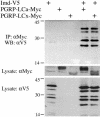
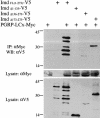
Drosophila peptidoglycan recognition protein LC (PGRP-LC), a transmembrane protein required for the response to bacterial infection, acts at the top of a cytoplasmic signaling cascade that requires the death-domain protein Imd and an IkappaB kinase to activate Relish, an NF-kappaB family member. It is not clear how binding of peptidoglycan to the extracellular domain of PGRP-LC activates intracellular signaling because its cytoplasmic domain has no homology to characterized proteins. Here, we demonstrate that PGRP-LC binds Imd and that its cytoplasmic domain is critical for its activity, suggesting that PGRP-LC acts as a signal-transducing receptor. The PGRP-LC cytoplasmic domain is also essential for the formation of dimers, and results suggest that dimerization may be required for receptor activation. The PGRP-LC cytoplasmic domain can mediate formation of heterodimers between different PGRP-LC isoforms, thereby potentially expanding the diversity of ligands that can be recognized by the receptor.
Figures
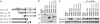
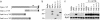

References
-
- Janeway, C. A., Jr., (1989) Cold Spring Harbor Symp. Quant. Biol. 54, 1–13. - PubMed
-
- Lemaitre, B., Nicolas, E., Michaut, L., Reichhart, J. M. & Hoffmann, J. A. (1996) Cell 86, 973–983. - PubMed
-
- Rutschmann, S., Kilinc, A. & Ferrandon, D. (2002) J. Immunol. 168, 1542–1546. - PubMed
-
- Levashina, E. A., Langley, E., Green, C., Gubb, D., Ashburner, M., Hoffmann, J. A. & Reichhart, J. M. (1999) Science 285, 1917–1919. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

