Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors
- PMID: 15657291
- PMCID: PMC2212785
- DOI: 10.1084/jem.20041535
Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors
Abstract
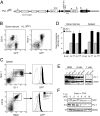
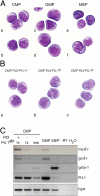
PU.1 is an Ets family transcription factor that is essential for fetal liver hematopoiesis. We have generated a PU.1(gfp) reporter strain that allowed us to examine the expression of PU.1 in all hematopoietic cell lineages and their early progenitors. Within the bone marrow progenitor compartment, PU.1 is highly expressed in the hematopoietic stem cell, the common lymphoid progenitor, and a proportion of common myeloid progenitors (CMPs). Based on Flt3 and PU.1 expression, the CMP could be divided into three subpopulations, Flt3(+) PU.1(hi), Flt3(-) PU.1(hi), and Flt3(-) PU.1(lo) CMPs. Colony-forming assays and in vivo lineage reconstitution demonstrated that the Flt3(+) PU.1(hi) and Flt3(-) PU.1(hi) CMPs were efficient precursors for granulocyte/macrophage progenitors (GMPs), whereas the Flt3(-) PU.1(lo) CMPs were highly enriched for committed megakaryocyte/erythrocyte progenitors (MEPs). CMPs have been shown to rapidly differentiate into GMPs and MEPs in vitro. Interestingly, short-term culture revealed that the Flt3(+) PU.1(hi) and Flt3(-) PU.1(hi) CMPs rapidly became CD16/32(high) (reminiscent of GMPs) in culture, whereas the Flt3(-) PU.1(lo) CMPs were the immediate precursors of the MEP. Thus, down-regulation of PU.1 expression in the CMP is the first molecularly identified event associated with the restriction of differentiation to erythroid and megakaryocyte lineages.
Figures


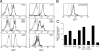

References
-
- Kondo, M., A.J. Wagers, M.G. Manz, S.S. Prohaska, D.C. Scherer, G.F. Beilhack, J.A. Shizuru, and I.L. Weissman. 2003. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu. Rev. Immunol. 21:759–806. - PubMed
-
- Akashi, K., D. Traver, T. Miyamoto, and I.L. Weissman. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197. - PubMed
-
- Cantor, A.B., and S.H. Orkin. 2002. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 21:3368–3376. - PubMed
-
- Warren, L.A., and E.V. Rothenberg. 2003. Regulatory coding of lymphoid lineage choice by hematopoietic transcription factors. Curr. Opin. Immunol. 15:166–175. - PubMed
-
- Scott, E.W., M.C. Simon, J. Anastasi, and H. Singh. 1994. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 265:1573–1577. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

