Rejoining of DNA double-strand breaks as a function of overhang length
- PMID: 15657419
- PMCID: PMC544009
- DOI: 10.1128/MCB.25.3.896-906.2005
Rejoining of DNA double-strand breaks as a function of overhang length
Abstract
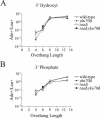
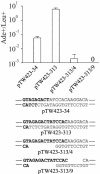
The ends of spontaneously occurring double-strand breaks (DSBs) may contain various lengths of single-stranded DNA, blocking lesions, and gaps and flaps generated by end annealing. To investigate the processing of such structures, we developed an assay in which annealed oligonucleotides are ligated onto the ends of a linearized plasmid which is then transformed into Saccharomyces cerevisiae. Reconstitution of a marker occurs only when the oligonucleotides are incorporated and repair is in frame, permitting rapid analysis of complex DSB ends. Here, we created DSBs with compatible overhangs of various lengths and asked which pathways are required for their precise repair. Three mechanisms of rejoining were observed, regardless of overhang polarity: nonhomologous end joining (NHEJ), a Rad52-dependent single-strand annealing-like pathway, and a third mechanism independent of the first two mechanisms. DSBs with overhangs of less than 4 bases were mainly repaired by NHEJ. Repair became less dependent on NHEJ when the overhangs were longer or had a higher GC content. Repair of overhangs greater than 8 nucleotides was as much as 150-fold more efficient, impaired 10-fold by rad52 mutation, and highly accurate. Reducing the microhomology extent between long overhangs reduced their repair dramatically, to less than NHEJ of comparable short overhangs. These data support a model in which annealing energy is a primary determinant of the rejoining efficiency and mechanism.
Figures




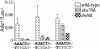

References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. E. Seidman, J. A. Smith, and K. Struhl (ed.). 1997. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
-
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
