New insights into DNA triplexes: residual twist and radial difference as measures of base triplet non-isomorphism and their implication to sequence-dependent non-uniform DNA triplex
- PMID: 15657986
- PMCID: PMC546132
- DOI: 10.1093/nar/gki143
New insights into DNA triplexes: residual twist and radial difference as measures of base triplet non-isomorphism and their implication to sequence-dependent non-uniform DNA triplex
Abstract
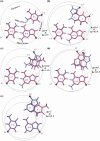
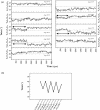
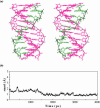
DNA triplexes are formed by both isomorphic (structurally alike) and non-isomorphic (structurally dissimilar) base triplets. It is espoused here that (i) the base triplet non-isomorphism may be articulated in structural terms by a residual twist (Delta(t) degrees), the angle formed by line joining the C1'...C1' atoms of the adjacent Hoogsteen or reverse Hoogsteen (RH) base pairs and the difference in base triplet radius (Delta(r) A), and (ii) their influence on DNA triplex is largely mechanistic, leading to the prediction of a high (t + Delta(t))degrees and low (t - Deltat)degrees twist at the successive steps of Hoogsteen or RH duplex of a parallel or antiparallel triplex. Efficacy of this concept is corroborated by molecular dynamics (MD) simulation of an antiparallel DNA triplex comprising alternating non-isomorphic G*GC and T*AT triplets. Conformational changes necessitated by base triplet non-isomorphism are found to induce an alternating (i) high anti and anti glycosyl and (ii) BII and an unusual BIII conformation resulting in a zigzag backbone for the RH strand. Thus, base triplet non-isomorphism causes DNA triplexes into exhibiting sequence-dependent non-uniform conformation. Such structural variations may be relevant in deciphering the specificity of interaction with DNA triplex binding proteins. Seemingly then, residual twist (Delta(t) degrees) and radial difference (Deltar A) suffice as indices to define and monitor the effect of base triplet non-isomorphism in nucleic acid triplexes.
Figures

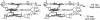



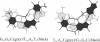

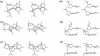
References
-
- Soyfer V.N., Potaman V.N. Triple-Helical Nucleic Acids. New York: Springer-Verlag; 1996.
-
- Praseuth D., Guieysse A.L., Helene C. Triple helix formation and the antigene strategy for sequence-specific control of gene expression. Biochim. Biophys. Acta. 1999;1489:181–206. - PubMed
-
- Wang E., Feigon J. Structures of nucleic acid triplexes. In: Neidle S., editor. Oxford Handbook of Nucleic Acid Structure. New York: Oxford University Press; 1999. pp. 355–388.
-
- Vasquez K.M., Glazer P.M. Triplex-forming oligonucleotides: principles and applications. Q. Rev. Biophys. 2002;35:89–107. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

