Separation of native prion protein (PrP) glycoforms by copper-binding using immobilized metal affinity chromatography (IMAC)
- PMID: 15658935
- PMCID: PMC1186727
- DOI: 10.1042/BJ20041291
Separation of native prion protein (PrP) glycoforms by copper-binding using immobilized metal affinity chromatography (IMAC)
Abstract
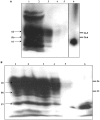
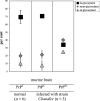
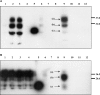
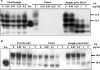
The conformational conversion of the normal cellular prion protein (PrPC) into the pathology-associated PrPSc isoform is a key event in TSEs (transmissible spongiform encephalopathies). The host PrPC molecule contains two N-linked glycosylation sites and binds copper under physiological conditions. In contrast with PrPC, PrPSc is insoluble in non-ionic detergents and does not bind to Cu2+ ions. Hence, we utilized copper binding to separate and characterize both PrP isoforms. Infected and uninfected murine brain and bovine stem brain specimens were treated with the mild non-ionic detergent n-octyl-beta-D-glucopyranoside (octylglucoside) to maintain the native PrP conformations during isolation. The solubilized homogenates were loaded on to Cu2+-saturated IMAC (immobilized metal affinity chromatography) columns and eluted using the chelating agent EDTA. Fractions were separated by SDS/PAGE and analysed by immunoblotting using anti-PrP monoclonal antibodies for glycosylation profiling. Whereas native PrPC and denatured PrPSc were retained by a Cu2+-loaded resin, native PrPSc and PrPres [PK (proteinase K)-resistant PrP] passed through the column. We demonstrate here that the IMAC technique is appropriate to isolate and partially purify PrPC from healthy brains in its native-like and biologically relevant glycosylated copper-binding forms. The IMAC technique is also well suited for the separation of native PrPC from aggregated PrPSc in infected brains. Our results indicate that in contrast with PrPSc in uninfected as well as infected brains, PrPC is predominantly present in the glycosylated forms.
Figures


Similar articles
-
Detection of bovine spongiform encephalopathy, ovine scrapie prion-related protein (PrPSc) and normal PrPc by monoclonal antibodies raised to copper-refolded prion protein.Biochem J. 2003 Feb 15;370(Pt 1):81-90. doi: 10.1042/BJ20021280. Biochem J. 2003. PMID: 12429022 Free PMC article.
-
Detection, quantification, and glycotyping of prion protein in specifically activated enzyme-linked immunosorbent assay plates.Anal Biochem. 2006 Dec 15;359(2):176-82. doi: 10.1016/j.ab.2006.10.002. Epub 2006 Oct 25. Anal Biochem. 2006. PMID: 17092479
-
Mapping of possible prion protein self-interaction domains using peptide arrays.BMC Biochem. 2007 Apr 12;8:6. doi: 10.1186/1471-2091-8-6. BMC Biochem. 2007. PMID: 17430579 Free PMC article.
-
Immunoaffinity purification and neutralization of scrapie prions.Prog Clin Biol Res. 1989;317:583-600. Prog Clin Biol Res. 1989. PMID: 2574871 Review.
-
Prion protein self-interactions: a gateway to novel therapeutic strategies?Vaccine. 2010 Nov 16;28(49):7810-23. doi: 10.1016/j.vaccine.2010.09.012. Epub 2010 Oct 20. Vaccine. 2010. PMID: 20932496 Review.
Cited by
-
Structural Consequences of Copper Binding to the Prion Protein.Cells. 2019 Jul 25;8(8):770. doi: 10.3390/cells8080770. Cells. 2019. PMID: 31349611 Free PMC article. Review.
-
Endogenous proteolytic cleavage of disease-associated prion protein to produce C2 fragments is strongly cell- and tissue-dependent.J Biol Chem. 2010 Apr 2;285(14):10252-64. doi: 10.1074/jbc.M109.083857. Epub 2010 Feb 12. J Biol Chem. 2010. PMID: 20154089 Free PMC article.
References
-
- McKinley M. P., Taraboulos A., Kenaga L., Serban D., Stieber A., DeArmond S. J., Prusiner S. B., Gonatas N. Ultrastructural localization of scrapie prion proteins in cytoplasmic vesicles of infected cultured cells. Lab. Invest. 1991;65:622–630. - PubMed
-
- Wadsworth J. D., Jackson G. S., Hill A. F., Collinge J. Molecular biology of prion propagation. Curr. Opin. Genet. Dev. 1999a;9:338–345. - PubMed
-
- MacGregor I. Prion protein and developments in its detection. Transfus. Med. 2001;11:3–14. - PubMed
-
- Martins V. R., Linden R., Prado M. A., Walz R., Sakamoto A. C., Izquierdo I., Brentani R. R. Cellular prion protein: on the road for functions. FEBS Lett. 2002;512:25–28. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

