Bicistronic and fused monocistronic transcripts are derived from adjacent loci in the Arabidopsis genome
- PMID: 15659355
- PMCID: PMC1370702
- DOI: 10.1261/rna.7114505
Bicistronic and fused monocistronic transcripts are derived from adjacent loci in the Arabidopsis genome
Abstract
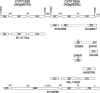
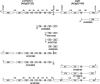
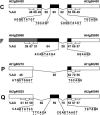
Comparisons of full-length cDNAs and genomic DNAs available for Arabidopsis thaliana described here indicate that some adjacent loci are transcribed into extremely long RNAs spanning two annotated genes. Once expressed, some of these transcripts are post-transcriptionally spliced within their coding and intergenic sequences to generate bicistronic transcripts containing two complete open reading frames. Others are spliced to generate monocistronic transcripts coding for fusion proteins with sequences derived from both loci. RT-PCR of several P450 transcripts in this collection indicates that these extended transcripts exist side by side with shorter monocistronic transcripts derived from the individual loci in each pair. The existence of these unusual transcripts highlights variations in the processes of transcription and splicing that could not possibly have been predicted in the algorithms used for genome annotation and splice site predictions.
Figures

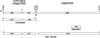




References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. - PubMed
-
- Blumenthal, T. 1995. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 11: 132–136. - PubMed
-
- ———. 1998. Gene clusters and polycistronic transcription in eukaryotes. BioEssays. 20: 480–487. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
