Stability of HAMLET--a kinetically trapped alpha-lactalbumin oleic acid complex
- PMID: 15659367
- PMCID: PMC2253409
- DOI: 10.1110/ps.04982905
Stability of HAMLET--a kinetically trapped alpha-lactalbumin oleic acid complex
Abstract
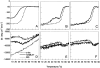
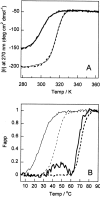
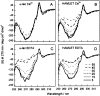
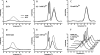
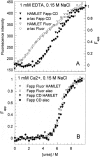
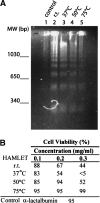
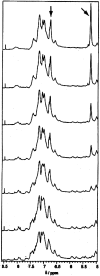
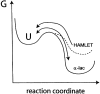
The stability toward thermal and urea denaturation was measured for HAMLET (human alpha-lactalbumin made lethal to tumor cells) and alpha-lactalbumin, using circular dichroism and fluorescence spectroscopy as well as differential scanning calorimetry. Under all conditions examined, HAMLET appears to have the same or lower stability than alpha-lactalbumin. The largest difference is seen for thermal denaturation of the calcium free (apo) forms, where the temperature at the transition midpoint is 15 degrees C lower for apo HAMLET than for apo alpha-lactalbumin. The difference becomes progressively smaller as the calcium concentration increases. Denaturation of HAMLET was found to be irreversible. Samples of HAMLET that have been renatured after denaturation have lost the specific biological activity toward tumor cells. Three lines of evidence indicate that HAMLET is a kinetic trap: (1) It has lower stability than alpha-lactalbumin, although it is a complex of alpha-lactalbumin and oleic acid; (2) its denaturation is irreversible and HAMLET is lost after denaturation; (3) formation of HAMLET requires a specific conversion protocol.
Figures

Similar articles
-
Who is Mr. HAMLET? Interaction of human alpha-lactalbumin with monomeric oleic acid.Biochemistry. 2008 Dec 9;47(49):13127-37. doi: 10.1021/bi801423s. Biochemistry. 2008. PMID: 19006329
-
Molten globule of bovine alpha-lactalbumin at neutral pH induced by heat, trifluoroethanol, and oleic acid: a comparative analysis by circular dichroism spectroscopy and limited proteolysis.Proteins. 2002 Nov 15;49(3):385-97. doi: 10.1002/prot.10234. Proteins. 2002. PMID: 12360528
-
No need to be HAMLET or BAMLET to interact with histones: binding of monomeric alpha-lactalbumin to histones and basic poly-amino acids.Biochemistry. 2004 May 18;43(19):5575-82. doi: 10.1021/bi049584y. Biochemistry. 2004. PMID: 15134431
-
Structure and function of human α-lactalbumin made lethal to tumor cells (HAMLET)-type complexes.FEBS J. 2010 Nov;277(22):4614-25. doi: 10.1111/j.1742-4658.2010.07890.x. FEBS J. 2010. PMID: 20977665 Review.
-
Can misfolded proteins be beneficial? The HAMLET case.Ann Med. 2009;41(3):162-76. doi: 10.1080/07853890802502614. Ann Med. 2009. PMID: 18985467 Review.
Cited by
-
Nm23-H1/NDP kinase folding intermediates and cancer: a hypothesis.J Bioenerg Biomembr. 2006 Aug;38(3-4):265-8. doi: 10.1007/s10863-006-9042-1. Epub 2006 Sep 1. J Bioenerg Biomembr. 2006. PMID: 16944300
-
Oleic acid may be the key contributor in the BAMLET-induced erythrocyte hemolysis and tumoricidal action.PLoS One. 2013 Sep 11;8(9):e68390. doi: 10.1371/journal.pone.0068390. eCollection 2013. PLoS One. 2013. PMID: 24039698 Free PMC article.
-
Drug-loaded oleic-acid grafted mesoporous silica nanoparticles conjugated with α-lactalbumin resembling BAMLET-like anticancer agent with improved biocompatibility and therapeutic efficacy.Mater Today Bio. 2022 May 4;15:100272. doi: 10.1016/j.mtbio.2022.100272. eCollection 2022 Jun. Mater Today Bio. 2022. PMID: 35607417 Free PMC article.
-
Protein receptor-independent plasma membrane remodeling by HAMLET: a tumoricidal protein-lipid complex.Sci Rep. 2015 Nov 12;5:16432. doi: 10.1038/srep16432. Sci Rep. 2015. PMID: 26561036 Free PMC article.
-
Free Fatty Acid and α-Lactalbumin-Oleic Acid Complexes in Preterm Human Milk Are Cytotoxic to Fetal Intestinal Cells in vitro.Front Nutr. 2022 Jul 5;9:918872. doi: 10.3389/fnut.2022.918872. eCollection 2022. Front Nutr. 2022. PMID: 35866080 Free PMC article.
References
-
- Acharya, K.R., Stuart, D.I., Walker, N.P., Lewis, M., and Phillips, D.C. 1989. Refined structure of baboon α-lactalbumin at 1.7 Å resolution. Comparison with c-type lysozyme. J. Mol. Biol. 208 99–127. - PubMed
-
- Acharya, K.R., Ren, J.S., Stuart, D.I., Phillips, D.C., and Fenna, R.E. 1991. Crystal structure of human α-lactalbumin at 1.7 Å resolution. J. Mol. Biol. 221 571–581. - PubMed
-
- Baker, D. and Agard, D.A. 1994. Kinetics versus thermodynamics in protein folding. Biochemistry 33 7505–7509. - PubMed
-
- Baldwin, T.O., Ziegler, M.M., Chaffotte, A.F., and Goldberg, M.E. 1993. Contribution of folding steps involving the individual subunits of bacterial luciferase to the assembly of the active heterodimeric enzyme. J. Biol. Chem. 268 10766–10772. - PubMed
-
- Brew, K. and Hill, R.L. 1975. Lactose biosynthesis. Rev. Physiol. Biochem. Pharmacol. 72 105–158. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

