SNP identification in unamplified human genomic DNA with gold nanoparticle probes
- PMID: 15659576
- PMCID: PMC548375
- DOI: 10.1093/nar/gni017
SNP identification in unamplified human genomic DNA with gold nanoparticle probes
Abstract
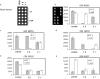
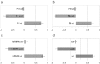
Single nucleotide polymorphisms (SNPs) comprise the most abundant source of genetic variation in the human genome. SNPs may be linked to genetic predispositions, frank disorders or adverse drug responses, or they may serve as genetic markers in linkage disequilibrium analysis. Thus far, established SNP detection techniques have utilized enzymes to meet the sensitivity and specificity requirements needed to overcome the high complexity of the human genome. Herein, we present for the first time a microarray-based method that allows multiplex SNP genotyping in total human genomic DNA without the need for target amplification or complexity reduction. This direct SNP genotyping methodology requires no enzymes and relies on the high sensitivity of the gold nanoparticle probes. Specificity is derived from two sequential oligonucleotide hybridizations to the target by allele-specific surface-immobilized capture probes and gene-specific oligonucleotide-functionalized gold nanoparticle probes. Reproducible multiplex SNP detection is demonstrated with unamplified human genomic DNA samples representing all possible genotypes for three genes involved in thrombotic disorders. The assay format is simple, rapid and robust pointing to its suitability for multiplex SNP profiling at the 'point of care'.
Figures



References
-
- Sachidanandam R. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. - PubMed
-
- Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. - PubMed
-
- Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. - PubMed
-
- Ardlie K.G., Kruglyak L., Seielstad M. Patterns of linkage disequilibrium in the human genome. Nature Rev. Genet. 2002;3:299–309. - PubMed
-
- Wall J.D., Pritchard J.K. Haplotype blocks and linkage disequilibrium in the human genome. Nature Rev. Genet. 2003;4:587–597. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

