Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid
- PMID: 15659623
- PMCID: PMC548830
- DOI: 10.1105/tpc.104.026690
Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid
Abstract
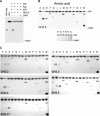
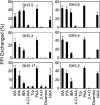
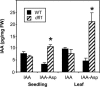
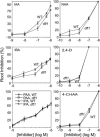
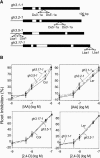
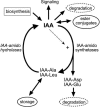
Substantial evidence indicates that amino acid conjugates of indole-3-acetic acid (IAA) function in auxin homeostasis, yet the plant enzymes involved in their biosynthesis have not been identified. We tested whether several Arabidopsis thaliana enzymes that are related to the auxin-induced soybean (Glycine max) GH3 gene product synthesize IAA-amino acid conjugates. In vitro reactions with six recombinant GH3 enzymes produced IAA conjugates with several amino acids, based on thin layer chromatography. The identity of the Ala, Asp, Phe, and Trp conjugates was verified by gas chromatography-mass spectrometry. Insertional mutations in GH3.1, GH3.2, GH3.5, and GH3.17 resulted in modestly increased sensitivity to IAA in seedling root. Overexpression of GH3.6 in the activation-tagged mutant dfl1-D did not significantly alter IAA level but resulted in 3.2- and 4.5-fold more IAA-Asp than in wild-type seedlings and mature leaves, respectively. In addition to IAA, dfl1-D was less sensitive to indole-3-butyric acid and naphthaleneacetic acid, consistent with the fact that GH3.6 was active on each of these auxins. By contrast, GH3.6 and the other five enzymes tested were inactive on halogenated auxins, and dfl1-D was not resistant to these. This evidence establishes that several GH3 genes encode IAA-amido synthetases, which help to maintain auxin homeostasis by conjugating excess IAA to amino acids.
Figures
Similar articles
-
Preference of Arabidopsis thaliana GH3.5 acyl amido synthetase for growth versus defense hormone acyl substrates is dictated by concentration of amino acid substrate aspartate.Phytochemistry. 2017 Nov;143:19-28. doi: 10.1016/j.phytochem.2017.07.001. Epub 2017 Jul 23. Phytochemistry. 2017. PMID: 28743075
-
Arabidopsis thaliana GH3.9 influences primary root growth.Planta. 2007 Jun;226(1):21-34. doi: 10.1007/s00425-006-0462-2. Epub 2007 Jan 11. Planta. 2007. PMID: 17216483
-
Arabidopsis thaliana GH3.5 acyl acid amido synthetase mediates metabolic crosstalk in auxin and salicylic acid homeostasis.Proc Natl Acad Sci U S A. 2016 Nov 29;113(48):13917-13922. doi: 10.1073/pnas.1612635113. Epub 2016 Nov 14. Proc Natl Acad Sci U S A. 2016. PMID: 27849615 Free PMC article.
-
Auxin: regulation, action, and interaction.Ann Bot. 2005 Apr;95(5):707-35. doi: 10.1093/aob/mci083. Epub 2005 Mar 4. Ann Bot. 2005. PMID: 15749753 Free PMC article. Review.
-
Connecting primary and specialized metabolism: Amino acid conjugation of phytohormones by GRETCHEN HAGEN 3 (GH3) acyl acid amido synthetases.Curr Opin Plant Biol. 2022 Apr;66:102194. doi: 10.1016/j.pbi.2022.102194. Epub 2022 Feb 23. Curr Opin Plant Biol. 2022. PMID: 35219141 Review.
Cited by
-
2,4-D and IAA Amino Acid Conjugates Show Distinct Metabolism in Arabidopsis.PLoS One. 2016 Jul 19;11(7):e0159269. doi: 10.1371/journal.pone.0159269. eCollection 2016. PLoS One. 2016. PMID: 27434212 Free PMC article.
-
Plant Hormone Homeostasis, Signaling, and Function during Adventitious Root Formation in Cuttings.Front Plant Sci. 2016 Mar 31;7:381. doi: 10.3389/fpls.2016.00381. eCollection 2016. Front Plant Sci. 2016. PMID: 27064322 Free PMC article. Review.
-
Integrated Signals of Jasmonates, Sugars, Cytokinins and Auxin Influence the Initial Growth of the Second Buds of Chrysanthemum after Decapitation.Biology (Basel). 2021 May 16;10(5):440. doi: 10.3390/biology10050440. Biology (Basel). 2021. PMID: 34065759 Free PMC article.
-
Distinct Characteristics of Indole-3-Acetic Acid and Phenylacetic Acid, Two Common Auxins in Plants.Plant Cell Physiol. 2015 Aug;56(8):1641-54. doi: 10.1093/pcp/pcv088. Epub 2015 Jun 14. Plant Cell Physiol. 2015. PMID: 26076971 Free PMC article.
-
Metabolomic, transcriptional, hormonal, and signaling cross-talk in superroot2.Mol Plant. 2010 Jan;3(1):192-211. doi: 10.1093/mp/ssp098. Epub 2009 Dec 14. Mol Plant. 2010. PMID: 20008451 Free PMC article.
References
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. - PubMed
-
- Bandurski, R.S., Cohen, J.D., Slovin, J.P., and Reinecke, D.M. (1995). Auxin biosynthesis and metabolism. In Plant Hormones, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 118–139.
-
- Bartel, B. (1997). Auxin biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 51–66. - PubMed
-
- Bartel, B., and Fink, G. (1995). ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268, 1745–1748. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

