The role of KNOX genes in the evolution of morphological novelty in Streptocarpus
- PMID: 15659624
- PMCID: PMC548817
- DOI: 10.1105/tpc.104.028936
The role of KNOX genes in the evolution of morphological novelty in Streptocarpus
Abstract
The genus Streptocarpus comprises species with diverse body plans. Caulescent species produce leaves from a conventional shoot apical meristem (SAM), whereas acaulescent species lack a conventional SAM and produce only a single leaf (the unifoliate form) or clusters of leaves from the base of more mature leaves (the rosulate form). These distinct morphologies reflect fundamental differences in the role of the SAM and the process of leaf specification. A subfamily of KNOTTED-like homeobox (KNOX) genes are known to be important in regulating meristem function and leaf development in model species with conventional morphologies. To test the involvement of KNOX genes in Streptocarpus evolution, two parologous KNOX genes (SSTM1 and SSTM2) were isolated from species with different growth forms. Their phylogenetic analysis suggested a gene duplication before the subgeneric split of Streptocarpus and resolved species relationships, supporting multiple evolutionary origins of the rosulate and unifoliate morphologies. In S. saxorum, a caulescent species with a conventional SAM, KNOX proteins were expressed in the SAM and transiently downregulated in incipient leaf primordia. The ability of acaulescent species to initiate leaves from existing leaves was found to correlate with SSTM1 expression and KNOX protein accumulation in leaves and to reflect genetic differences at two loci. Neither locus corresponded to SSTM1, suggesting that cis-acting differences in SSTM1 regulation were not responsible for evolution of the rosulate and unifoliate forms. However, the involvement of KNOX proteins in leaf formation in rosulate species suggests that they have played an indirect role in the development of morphological diversity in Streptocarpus.
Figures





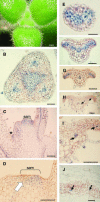
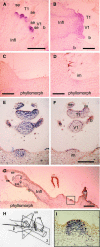

References
-
- Barton, M.K., and Poethig, R.S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of development in the wild type and in the shoot meristemless mutant. Development 119, 823–831.
-
- Bharathan, G., Goliber, T.E., Moore, C., Kessler, S., Pham, T., and Sinha, N.R. (2002). Homologies in leaf form inferred from KNOX1 gene expression during development. Science 296, 1858–1860. - PubMed
-
- Bürglin, T.R. (1998). The PBC domain contains a MEINOX domain: Coevolution of Hox and TALE homeobox genes? Dev. Genes Evol. 208, 113–116. - PubMed
-
- Burtt, B.L. (1970). Studies in the Gesneriaceae of the Old World XXXI: Some aspects of functional evolution. Notes R. Bot. Gard. (Edinb.) 30, 1–10.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

