Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19
- PMID: 15659631
- PMCID: PMC548818
- DOI: 10.1105/tpc.104.028316
Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19
Abstract
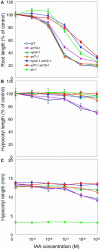
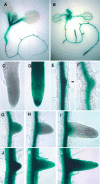
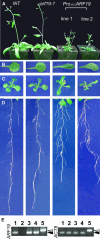
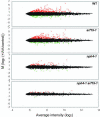
The AUXIN RESPONSE FACTOR (ARF) gene family products, together with the AUXIN/INDOLE-3-ACETIC ACID proteins, regulate auxin-mediated transcriptional activation/repression. The biological function(s) of most ARFs is poorly understood. Here, we report the identification and characterization of T-DNA insertion lines for 18 of the 23 ARF gene family members in Arabidopsis thaliana. Most of the lines fail to show an obvious growth phenotype except of the previously identified arf2/hss, arf3/ett, arf5/mp, and arf7/nph4 mutants, suggesting that there are functional redundancies among the ARF proteins. Subsequently, we generated double mutants. arf7 arf19 has a strong auxin-related phenotype not observed in the arf7 and arf19 single mutants, including severely impaired lateral root formation and abnormal gravitropism in both hypocotyl and root. Global gene expression analysis revealed that auxin-induced gene expression is severely impaired in the arf7 single and arf7 arf19 double mutants. For example, the expression of several genes, such as those encoding members of LATERAL ORGAN BOUNDARIES domain proteins and AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE, are disrupted in the double mutant. The data suggest that the ARF7 and ARF19 proteins play essential roles in auxin-mediated plant development by regulating both unique and partially overlapping sets of target genes. These observations provide molecular insight into the unique and overlapping functions of ARF gene family members in Arabidopsis.
Figures





Similar articles
-
The IAA7-ARF7-ARF19 auxin signaling module plays diverse roles in Arabidopsis growth and development.Planta. 2025 Jun 3;262(1):12. doi: 10.1007/s00425-025-04731-z. Planta. 2025. PMID: 40459652
-
ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis.Plant Cell. 2007 Jan;19(1):118-30. doi: 10.1105/tpc.106.047761. Epub 2007 Jan 26. Plant Cell. 2007. PMID: 17259263 Free PMC article.
-
MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana.Plant Cell. 2004 Feb;16(2):379-93. doi: 10.1105/tpc.018630. Epub 2004 Jan 16. Plant Cell. 2004. PMID: 14729917 Free PMC article.
-
Game of thrones among AUXIN RESPONSE FACTORs-over 30 years of MONOPTEROS research.J Exp Bot. 2023 Dec 1;74(22):6904-6921. doi: 10.1093/jxb/erad272. J Exp Bot. 2023. PMID: 37450945 Free PMC article. Review.
-
Auxin response factors are keys to the many auxin doors.New Phytol. 2022 Jul;235(2):402-419. doi: 10.1111/nph.18159. Epub 2022 May 10. New Phytol. 2022. PMID: 35434800 Review.
Cited by
-
Overexpression of TaLBD16-4D alters plant architecture and heading date in transgenic wheat.Front Plant Sci. 2022 Sep 21;13:911993. doi: 10.3389/fpls.2022.911993. eCollection 2022. Front Plant Sci. 2022. PMID: 36212357 Free PMC article.
-
Transcription Factors and Their Regulatory Roles in the Male Gametophyte Development of Flowering Plants.Int J Mol Sci. 2024 Jan 1;25(1):566. doi: 10.3390/ijms25010566. Int J Mol Sci. 2024. PMID: 38203741 Free PMC article. Review.
-
Comparative transcriptomics analysis of developing peanut (Arachis hypogaea L.) pods reveals candidate genes affecting peanut seed size.Front Plant Sci. 2022 Sep 12;13:958808. doi: 10.3389/fpls.2022.958808. eCollection 2022. Front Plant Sci. 2022. PMID: 36172561 Free PMC article.
-
Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors.Proc Natl Acad Sci U S A. 2012 Nov 27;109(48):19537-44. doi: 10.1073/pnas.1214774109. Epub 2012 Nov 9. Proc Natl Acad Sci U S A. 2012. PMID: 23144218 Free PMC article.
-
Interplay between sucrose and folate modulates auxin signaling in Arabidopsis.Plant Physiol. 2013 Jul;162(3):1552-65. doi: 10.1104/pp.113.215095. Epub 2013 May 20. Plant Physiol. 2013. PMID: 23690535 Free PMC article.
References
-
- Abel, S., Ballas, N., Wong, L.-M., and Theologis, A. (1996). DNA elements responsive to auxin. Bioessays 18, 647–654. - PubMed
-
- Abel, S., Nguyen, M.D., and Theologis, A. (1995). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251, 533–549. - PubMed
-
- Alonso, J.M.A., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

