Large intraspecific haplotype variability at the Rph7 locus results from rapid and recent divergence in the barley genome
- PMID: 15659632
- PMCID: PMC548812
- DOI: 10.1105/tpc.104.028225
Large intraspecific haplotype variability at the Rph7 locus results from rapid and recent divergence in the barley genome
Abstract
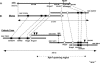
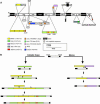
To study genome evolution and diversity in barley (Hordeum vulgare), we have sequenced and compared more than 300 kb of sequence spanning the Rph7 leaf rust disease resistance gene in two barley cultivars. Colinearity was restricted to five genic and two intergenic regions representing <35% of the two sequences. In each interval separating the seven conserved regions, the number and type of repetitive elements were completely different between the two homologous sequences, and a single gene was absent in one cultivar. In both cultivars, the nonconserved regions consisted of approximately 53% repetitive sequences mainly represented by long-terminal repeat retrotransposons that have inserted <1 million years ago. PCR-based analysis of intergenic regions at the Rph7 locus and at three other independent loci in 41 H. vulgare lines indicated large haplotype variability in the cultivated barley gene pool. Together, our data indicate rapid and recent divergence at homologous loci in the genome of H. vulgare, possibly providing the molecular mechanism for the generation of high diversity in the barley gene pool. Finally, comparative analysis of the gene composition in barley, wheat (Triticum aestivum), rice (Oryza sativa), and sorghum (Sorghum bicolor) suggested massive gene movements at the Rph7 locus in the Triticeae lineage.
Figures

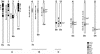
References
-
- Badr, A., Muller, K., Schafer-Pregl, R., El Rabey, H., Effgen, S., Ibrahim, H.H., Pozzi, C., Rohde, W., and Salamini, F. (2000). On the origin and domestication history of Barley (Hordeum vulgare). Mol. Biol. Evol. 17, 499–510. - PubMed
-
- Bennetzen, J.L. (2002). Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica 115, 29–36. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

