Plus and minus sexual agglutinins from Chlamydomonas reinhardtii
- PMID: 15659633
- PMCID: PMC548829
- DOI: 10.1105/tpc.104.028035
Plus and minus sexual agglutinins from Chlamydomonas reinhardtii
Abstract
Gametes of the unicellular green alga Chlamydomonas reinhardtii undergo sexual adhesion via enormous chimeric Hyp-rich glycoproteins (HRGPs), the plus and minus sexual agglutinins, that are displayed on their flagellar membrane surfaces. We have previously purified the agglutinins and analyzed their structural organization using electron microscopy. We report here the cloning and sequencing of the Sag1 and Sad1 genes that encode the two agglutinins and relate their derived amino acid sequences and predicted secondary structure to the morphology of the purified proteins. Both agglutinin proteins are organized into three distinct domains: a head, a shaft in a polyproline II configuration, and an N-terminal domain. The plus and minus heads are related in overall organization but poorly conserved in sequence except for two regions of predicted hydrophobic alpha-helix. The shafts contain numerous repeats of the PPSPX motif previously identified in Gp1, a cell wall HRGP. We propose that the head domains engage in autolectin associations with the distal termini of their own shafts and suggest ways that adhesion may involve head-head interactions, exolectin interactions between the heads and shafts of opposite type, and antiparallel shaft-shaft interactions mediated by carbohydrates displayed in polyproline II configurations.
Figures
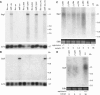








Similar articles
-
Between-species analysis of short-repeat modules in cell wall and sex-related hydroxyproline-rich glycoproteins of Chlamydomonas.Plant Physiol. 2007 Aug;144(4):1813-26. doi: 10.1104/pp.107.100891. Epub 2007 Jun 15. Plant Physiol. 2007. PMID: 17573538 Free PMC article.
-
Glycosylated polyproline II rods with kinks as a structural motif in plant hydroxyproline-rich glycoproteins.Biochemistry. 2001 Mar 6;40(9):2978-87. doi: 10.1021/bi0023605. Biochemistry. 2001. PMID: 11258910
-
A sex recognition glycoprotein is encoded by the plus mating-type gene fus1 of Chlamydomonas reinhardtii.Mol Biol Cell. 1996 Aug;7(8):1235-48. doi: 10.1091/mbc.7.8.1235. Mol Biol Cell. 1996. PMID: 8856667 Free PMC article.
-
Sex determination in Chlamydomonas.Semin Cell Dev Biol. 2007 Jun;18(3):350-61. doi: 10.1016/j.semcdb.2007.02.006. Epub 2007 Feb 25. Semin Cell Dev Biol. 2007. PMID: 17643326 Review.
-
Sexual reproduction and sex determination in green algae.J Plant Res. 2017 May;130(3):423-431. doi: 10.1007/s10265-017-0908-6. Epub 2017 Feb 10. J Plant Res. 2017. PMID: 28188480 Review.
Cited by
-
Dynamic Changes in the Transcriptome and Methylome of Chlamydomonas reinhardtii throughout Its Life Cycle.Plant Physiol. 2015 Dec;169(4):2730-43. doi: 10.1104/pp.15.00861. Epub 2015 Oct 8. Plant Physiol. 2015. PMID: 26450704 Free PMC article.
-
Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri.Science. 2010 Jul 9;329(5988):223-6. doi: 10.1126/science.1188800. Science. 2010. PMID: 20616280 Free PMC article.
-
Evolution of uni- and bifactorial sexual compatibility systems in fungi.Heredity (Edinb). 2013 Dec;111(6):445-55. doi: 10.1038/hdy.2013.67. Epub 2013 Jul 10. Heredity (Edinb). 2013. PMID: 23838688 Free PMC article. Review.
-
Uni-directional ciliary membrane protein trafficking by a cytoplasmic retrograde IFT motor and ciliary ectosome shedding.Elife. 2015 Feb 17;4:e05242. doi: 10.7554/eLife.05242. Elife. 2015. PMID: 25688564 Free PMC article.
-
Intrinsically Disordered Proteins: Critical Components of the Wetware.Chem Rev. 2022 Mar 23;122(6):6614-6633. doi: 10.1021/acs.chemrev.1c00848. Epub 2022 Feb 16. Chem Rev. 2022. PMID: 35170314 Free PMC article. Review.
References
-
- Adair, W.S. (1985). Characterization of Chlamydomonas sexual agglutinins. J. Cell Sci. 2 (suppl.), 233–260. - PubMed
-
- Adair, W.S., Hwang, C., and Goodenough, U.W. (1983). Identification and visualization of the sexual agglutinin from mating-type plus flagellar membranes of Chlamydomonas. Cell 33, 183–193. - PubMed
-
- Adair, W.S., Monk, B.C., Cohen, R., Hwang, C., and Goodenough, U.W. (1982). Sexual agglutinins from the Chlamydomonas flagellar membrane: Partial purification and characterization. J. Biol. Chem. 257, 4593–4602. - PubMed
-
- Armbrust, E.V., Ferris, P.J., and Goodenough, U.W. (1993). A mating type-linked gene cluster expressed in Chlamydomonas participates in the uniparental inheritance of the chloroplast genome. Cell 74, 801–811. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

