DISTORTED3/SCAR2 is a putative arabidopsis WAVE complex subunit that activates the Arp2/3 complex and is required for epidermal morphogenesis
- PMID: 15659634
- PMCID: PMC548822
- DOI: 10.1105/tpc.104.027987
DISTORTED3/SCAR2 is a putative arabidopsis WAVE complex subunit that activates the Arp2/3 complex and is required for epidermal morphogenesis
Abstract
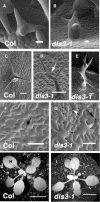
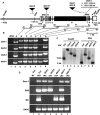
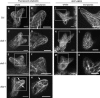
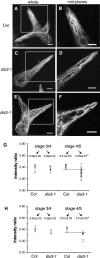
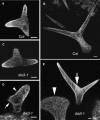
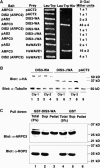
In a plant cell, a subset of actin filaments function as a scaffold that positions the endomembrane system and acts as a substrate on which organelle motility occurs. Other actin filament arrays appear to be more dynamic and reorganize in response to growth signals and external cues. The distorted group of trichome morphology mutants provides powerful genetic tools to study the control of actin filament nucleation in the context of morphogenesis. In this article, we report that DISTORTED3 (DIS3) encodes a plant-specific SCAR/WAVE homolog. Null alleles of DIS3, like those of other Arabidopsis thaliana WAVE and Actin-Related Protein (ARP) 2/3 subunit genes, cause trichome distortion, defects in cell-cell adhesion, and reduced hypocotyl growth in etiolated seedlings. DIS3 efficiently activates the actin filament nucleation and branching activity of vertebrate Arp2/3 and functions within a WAVE-ARP2/3 pathway in vivo. DIS3 may assemble into a WAVE complex via a physical interaction with a highly diverged Arabidopsis Abi-1-like bridging protein. These results demonstrate the utility of the Arabidopsis trichome system to understand how the WAVE and ARP2/3 complexes translate signaling inputs into a coordinated morphogenetic response.
Figures






References
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. - PubMed
-
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1994). Current protocols in molecular biology. (New York: John Wiley & Sons).
-
- Basu, D., El-Assal, S.E., Le, J., Mallery, E.L., and Szymanski, D.B. (2004). Interchangeable functions of Arabidopsis PIROGI and the human WAVE complex subunit SRA-1 during leaf epidermal morphogenesis. Development 131, 4345–4355. - PubMed
-
- Blagg, S.L., and Insall, R.H. (2004). Solving the WAVE function. Nat. Cell Biol. 6, 279–281. - PubMed
NOTE ADDED IN PROOF
-
- While this work was under review, Frank et al. (2004) reported that plant SCAR proteins bind to BRK1 (HSPC300) and can activate vertebrate Arp2/3.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

