Mx-rMx, a family of interacting transposons in the growing hAT superfamily of maize
- PMID: 15659635
- PMCID: PMC548813
- DOI: 10.1105/tpc.104.027797
Mx-rMx, a family of interacting transposons in the growing hAT superfamily of maize
Abstract
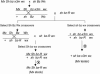
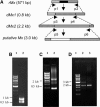
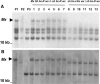
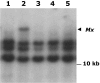
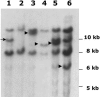
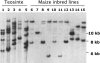
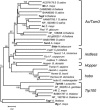
More than half a century after the discovery of transposable elements, the number of genetically defined autonomous elements that have been isolated and characterized molecularly in any one species remains surprisingly small. Because of its rich genetic history, maize (Zea mays) is, by far, the plant with the largest number of such elements. Yet, even in maize, a maximum of only two autonomous elements have been characterized in any transposon superfamily. This article describes the isolation and molecular and genetic characterization of Mx (for mobile element induced by x-rays), a third autonomous member of the hAT transposon superfamily in maize. Mx is 3731 bp long, ends in 13-bp terminal inverted repeats (TIRs), and causes an 8-bp duplication of the target site. Mx and rMx (for responder to Mx), its 571-bp nonautonomous partner, define a classical family of interacting transposable elements. Surprisingly, the TIRs of Mx and rMx are only 73% identical, and the subterminal sequences are even less so, suggesting that Mx and rMx may represent diverging transposable elements still capable of mobilization by the same transposase. Sequences that are closer to the ends of either Mx or rMx are present in the maize genome. Mx is predicted to encode a 674-amino acid protein that is homologous to the Ac transposase. Although Mx and Ac are closely related, they do not interact. Other data suggest that maize may possess at least five families of hAT transposons that do not interact with each other. The possible origin of noninteracting transposon families within the same superfamily is discussed.
Figures



Similar articles
-
The complete Ac/Ds transposon family of maize.BMC Genomics. 2011 Dec 1;12:588. doi: 10.1186/1471-2164-12-588. BMC Genomics. 2011. PMID: 22132901 Free PMC article.
-
Structure and evolution of the hAT transposon superfamily.Genetics. 2001 Jul;158(3):949-57. doi: 10.1093/genetics/158.3.949. Genetics. 2001. PMID: 11454746 Free PMC article.
-
Identification of a high frequency transposon induced by tissue culture, nDaiZ, a member of the hAT family in rice.Genomics. 2009 Mar;93(3):274-81. doi: 10.1016/j.ygeno.2008.11.007. Epub 2008 Dec 23. Genomics. 2009. PMID: 19071208
-
Molecular biology of maize Ac/Ds elements: an overview.Methods Mol Biol. 2013;1057:59-82. doi: 10.1007/978-1-62703-568-2_5. Methods Mol Biol. 2013. PMID: 23918421 Review.
-
McClintock's controlling elements: the full story.Cytogenet Genome Res. 2005;109(1-3):90-103. doi: 10.1159/000082387. Cytogenet Genome Res. 2005. PMID: 15753564 Review.
Cited by
-
Survey of sugar beet (Beta vulgaris L.) hAT transposons and MITE-like hATpin derivatives.Plant Mol Biol. 2012 Mar;78(4-5):393-405. doi: 10.1007/s11103-011-9872-z. Epub 2012 Jan 13. Plant Mol Biol. 2012. PMID: 22246381
-
A novel active transposon creates allelic variation through altered translation rate to influence protein abundance.Nucleic Acids Res. 2023 Jan 25;51(2):595-609. doi: 10.1093/nar/gkac1195. Nucleic Acids Res. 2023. PMID: 36629271 Free PMC article.
-
TCUP: A Novel hAT Transposon Active in Maize Tissue Culture.Front Plant Sci. 2012 Jan 26;3:6. doi: 10.3389/fpls.2012.00006. eCollection 2012. Front Plant Sci. 2012. PMID: 22639634 Free PMC article.
-
Scanning of transposable elements and analyzing expression of transposase genes of sweet potato [Ipomoea batatas].PLoS One. 2014 Mar 7;9(3):e90895. doi: 10.1371/journal.pone.0090895. eCollection 2014. PLoS One. 2014. PMID: 24608103 Free PMC article.
-
The diversification and activity of hAT transposons in Musa genomes.Chromosome Res. 2014 Dec;22(4):559-71. doi: 10.1007/s10577-014-9445-5. Epub 2014 Nov 7. Chromosome Res. 2014. PMID: 25377178
References
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Becker, H.A., and Kunze, R. (1996). Binding sites for maize nuclear proteins in the subterminal regions of the transposable element Activator. Mol. Gen. Genet. 251, 428–435. - PubMed
-
- Becker, H.A., and Kunze, R. (1997). Maize Activator transposase has a bipartite DNA binding domain that recognizes subterminal sequences and the terminal inverted repeats. Mol. Gen. Genet. 254, 219–230. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

