The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis
- PMID: 15659636
- PMCID: PMC548820
- DOI: 10.1105/tpc.104.027722
The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis
Abstract
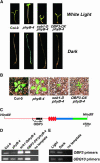
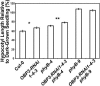
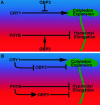
Plants perceive subtle changes in light quality and quantity through a set of photoreceptors, including phytochromes and cryptochromes. Upon perception, these photoreceptors initiate signal transduction pathways leading to photomorphogenic changes in development. Using activation-tagging mutagenesis to identify novel light-signaling components, we have isolated a gain-of-function mutant, sob1-D (suppressor of phytochrome B-4 [phyB-4] dominant), which suppresses the long-hypocotyl phenotype of the phyB missense allele, phyB-4. The sob1-D mutant phenotype is caused by the overexpression of a Dof (DNA binding with one finger) transcription factor, OBF4 Binding Protein 3 (OBP3). A translational fusion between OBP3 and green fluorescent protein is nuclear localized in onion (Allium cepa) cells. Tissue-specific accumulation of an OBP3:OBP3-beta-glucuronidase translational fusion is regulated by light in Arabidopsis thaliana. Hypocotyls of transgenic lines with reduced OBP3 expression are less responsive to red light. This aberrant phenotype in red light requires functional phyB, suggesting that OBP3 is a positive regulator of phyB-mediated inhibition of hypocotyl elongation. Furthermore, these partial-loss-of-function lines have larger cotyledons. This light-dependent cotyledon phenotype is most dramatic in blue light and requires functional cryptochrome 1 (cry1), indicating that OBP3 is a negative regulator of cry1-mediated cotyledon expansion. These results suggest a model where OBP3 is a component in both phyB and cry1 signaling pathways, acting as a positive and negative regulator, respectively. An alternate, though not mutually exclusive, model places OBP3 as a general inhibitor of tissue expansion with phyB and cry1, differentially modulating OBP3's role in this response.
Figures





Similar articles
-
Roles for the N- and C-terminal domains of phytochrome B in interactions between phytochrome B and cryptochrome signaling cascades.Plant Cell Physiol. 2007 Mar;48(3):424-33. doi: 10.1093/pcp/pcm012. Epub 2007 Jan 23. Plant Cell Physiol. 2007. PMID: 17251203
-
HYPERSENSITIVE TO RED AND BLUE 1, a ZZ-type zinc finger protein, regulates phytochrome B-mediated red and cryptochrome-mediated blue light responses.Plant Cell. 2005 Mar;17(3):822-35. doi: 10.1105/tpc.104.029165. Epub 2005 Feb 10. Plant Cell. 2005. PMID: 15705950 Free PMC article.
-
Photoexcited CRY1 and phyB interact directly with ARF6 and ARF8 to regulate their DNA-binding activity and auxin-induced hypocotyl elongation in Arabidopsis.New Phytol. 2020 Jan;225(2):848-865. doi: 10.1111/nph.16194. Epub 2019 Oct 25. New Phytol. 2020. PMID: 31514232
-
Signaling mechanisms of plant cryptochromes in Arabidopsis thaliana.J Plant Res. 2016 Mar;129(2):137-48. doi: 10.1007/s10265-015-0782-z. Epub 2016 Jan 25. J Plant Res. 2016. PMID: 26810763 Free PMC article. Review.
-
TANDEM ZINC-FINGER/PLUS3: a multifaceted integrator of light signaling.Trends Plant Sci. 2025 Jun;30(6):654-664. doi: 10.1016/j.tplants.2024.11.014. Epub 2024 Dec 18. Trends Plant Sci. 2025. PMID: 39701906 Review.
Cited by
-
Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor.Planta. 2015 Mar;241(3):549-62. doi: 10.1007/s00425-014-2239-3. Epub 2015 Jan 7. Planta. 2015. PMID: 25564353 Review.
-
Increasing the performance of cucumber (Cucumis sativus L.) seedlings by LED illumination.Sci Rep. 2022 Jan 17;12(1):852. doi: 10.1038/s41598-022-04859-y. Sci Rep. 2022. PMID: 35039577 Free PMC article.
-
The growth, nutrient uptake and fruit quality in four strawberry cultivars under different Spectra of LED supplemental light.BMC Plant Biol. 2024 Mar 8;24(1):179. doi: 10.1186/s12870-024-04880-5. BMC Plant Biol. 2024. PMID: 38454341 Free PMC article.
-
The Cryptochrome Blue Light Receptors.Arabidopsis Book. 2010 Sep 23;8:e0135. doi: 10.1199/tab.0135. Arabidopsis Book. 2010. PMID: 21841916 Free PMC article.
-
AtDOF5.4/OBP4, a DOF Transcription Factor Gene that Negatively Regulates Cell Cycle Progression and Cell Expansion in Arabidopsis thaliana.Sci Rep. 2016 Jun 14;6:27705. doi: 10.1038/srep27705. Sci Rep. 2016. PMID: 27297966 Free PMC article.
References
-
- Ahmad, M., and Cashmore, A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366, 162–166. - PubMed
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. - PubMed
-
- Bauer, D., Viczian, A., Kircher, S., Nobis, T., Nitschke, R., Kunkel, T., Panigrahi, K.C., Adam, E., Fejes, E., Schafer, E., and Nagy, F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16, 1433–1445. - PMC - PubMed
-
- Briggs, W.R., and Christie, J.M. (2002). Phototropins 1 and 2: Versatile plant blue-light receptors. Trends Plant Sci. 7, 204–210. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

