Genetic analysis reveals domain interactions of Arabidopsis Hsp100/ClpB and cooperation with the small heat shock protein chaperone system
- PMID: 15659638
- PMCID: PMC548826
- DOI: 10.1105/tpc.104.027540
Genetic analysis reveals domain interactions of Arabidopsis Hsp100/ClpB and cooperation with the small heat shock protein chaperone system
Abstract
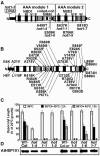
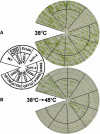
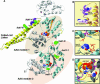
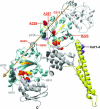
We have defined amino acids important for function of the Arabidopsis thaliana Hsp100/ClpB chaperone (AtHsp101) in acquired thermotolerance by isolating recessive, loss-of-function mutations and a novel semidominant, gain-of-function allele [hot1-4 (A499T)]. The hot1-4 allele is unusual in that it not only fails to develop thermotolerance to 45 degrees C after acclimation at 38 degrees C, but also is sensitive to 38 degrees C, which is a permissive temperature for wild-type and loss-of-function mutants. hot1-4 lies between nucleotide binding domain 1 (NBD1) and NBD2 in a coiled-coil domain that is characteristic of the Hsp100/ClpB proteins. We then isolated two classes of intragenic suppressor mutations of hot1-4: loss-of-function mutations (Class 1) that eliminated the 38 degrees C sensitivity, but did not restore thermotolerance function to hot1-4, and Class 2 suppressors that restored acquired thermotolerance function to hot1-4. Location of the hot1-4 Class 2 suppressors supports a functional link between the coiled-coil domain and both NBD1 and the axial channel of the Hsp100/ClpB hexamer. In addition, the strongest Class 2 suppressors restored solubility of aggregated small heat shock proteins (sHsps) after heat stress, revealing genetic interaction of the Hsp100/ClpB and sHsp chaperone systems. These results also demonstrate that quantitative phenotypes can be used for in vivo genetic dissection of protein mechanism in Arabidopsis.
Figures





Similar articles
-
The Arabidopsis ClpB/Hsp100 family of proteins: chaperones for stress and chloroplast development.Plant J. 2007 Jan;49(1):115-27. doi: 10.1111/j.1365-313X.2006.02940.x. Epub 2006 Nov 28. Plant J. 2007. PMID: 17144892
-
Crystal structure of E. coli Hsp100 ClpB nucleotide-binding domain 1 (NBD1) and mechanistic studies on ClpB ATPase activity.J Mol Biol. 2002 May 10;318(4):1127-37. doi: 10.1016/S0022-2836(02)00188-2. J Mol Biol. 2002. PMID: 12054807
-
Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress.Proc Natl Acad Sci U S A. 2000 Apr 11;97(8):4392-7. doi: 10.1073/pnas.97.8.4392. Proc Natl Acad Sci U S A. 2000. PMID: 10760305 Free PMC article.
-
ClpB/Hsp100 proteins and heat stress tolerance in plants.Crit Rev Biotechnol. 2016 Oct;36(5):862-74. doi: 10.3109/07388551.2015.1051942. Epub 2015 Jun 30. Crit Rev Biotechnol. 2016. PMID: 26121931 Review.
-
The ClpB/Hsp104 molecular chaperone-a protein disaggregating machine.J Struct Biol. 2004 Apr-May;146(1-2):99-105. doi: 10.1016/j.jsb.2003.11.016. J Struct Biol. 2004. PMID: 15037241 Review.
Cited by
-
Mutations in an Arabidopsis mitochondrial transcription termination factor-related protein enhance thermotolerance in the absence of the major molecular chaperone HSP101.Plant Cell. 2012 Aug;24(8):3349-65. doi: 10.1105/tpc.112.101006. Epub 2012 Aug 31. Plant Cell. 2012. PMID: 22942382 Free PMC article.
-
Silencing of class I small heat shock proteins affects seed-related attributes and thermotolerance in rice seedlings.Planta. 2019 Dec 3;251(1):26. doi: 10.1007/s00425-019-03318-9. Planta. 2019. PMID: 31797121
-
Mutant Analysis Reveals Allosteric Regulation of ClpB Disaggregase.Front Mol Biosci. 2017 Feb 22;4:6. doi: 10.3389/fmolb.2017.00006. eCollection 2017. Front Mol Biosci. 2017. PMID: 28275610 Free PMC article.
-
A tightly regulated molecular toggle controls AAA+ disaggregase.Nat Struct Mol Biol. 2012 Dec;19(12):1338-46. doi: 10.1038/nsmb.2441. Epub 2012 Nov 18. Nat Struct Mol Biol. 2012. PMID: 23160353
-
Enhancement of reproductive heat tolerance in plants.PLoS One. 2015 Apr 7;10(4):e0122933. doi: 10.1371/journal.pone.0122933. eCollection 2015. PLoS One. 2015. PMID: 25849955 Free PMC article.
References
-
- Beinker, P., Schlee, S., Groemping, Y., Seidel, R., and Reinstein, J. (2002). The N terminus of ClpB from Thermus thermophilus is not essential for the chaperone activity. J. Biol. Chem. 277, 47160–47166. - PubMed
-
- Cashikar, A.G., Schirmer, E.C., Hattendorf, D.A., Glover, J.R., Ramakrishnan, M.S., Ware, D.M., and Lindquist, S.L. (2002). Defining a pathway of communication from the C-terminal peptide binding domain to the N-terminal ATPase domain in a AAA protein. Mol. Cell 9, 751–760. - PubMed
NOTE ADDED IN PROOF
-
- Additional published data support threading of the substrate through the axial channel.
-
- Weibezahn, J., Tessarz, P., Schlieker, C., Zahn, R., Maglica, Z., Lee, S., Zentgraf, H., Weber-Ban, E.U., Dougan, D.A., Tsai, F.T.F., Mogk, A., and Bukau, B. (2004). Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119, 653–665. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

