The functionally conserved nucleoporins Nup124p from fission yeast and the human Nup153 mediate nuclear import and activity of the Tf1 retrotransposon and HIV-1 Vpr
- PMID: 15659641
- PMCID: PMC1073664
- DOI: 10.1091/mbc.e04-07-0583
The functionally conserved nucleoporins Nup124p from fission yeast and the human Nup153 mediate nuclear import and activity of the Tf1 retrotransposon and HIV-1 Vpr
Abstract
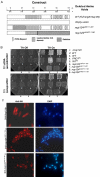
We report that the fission yeast nucleoporin Nup124p is required for the nuclear import of both, retrotransposon Tf1-Gag as well as the retroviral HIV-1 Vpr. Failure to import Tf1-Gag into the nucleus in a nup124 null mutant resulted in complete loss of Tf1 transposition. Similarly, nuclear import of HIV-1 Vpr was impaired in nup124 null mutant strains and cells became resistant to Vpr's cell-killing activity. On the basis of protein domain similarity, the human nucleoporin Nup153 was identified as a putative homolog of Nup124p. We demonstrate that in vitro-translated Nup124p and Nup153 coimmunoprecipitate Tf1-Gag or HIV-1 Vpr. Though full-length Nup153 was unable to complement the Tf1 transposition defect in a nup124 null mutant, we provide evidence that both nucleoporins share a unique N-terminal domain, Nup124p(AA264-454) and Nup153(AA448-634) that is absolutely essential for Tf1 transposition. Epigenetic overexpression of this domain in a wild-type (nup124(+)) background blocked Tf1 activity implying that sequences from Nup124p and the human Nup153 challenged the same pathway affecting Tf1 transposition. Our results establish a unique relationship between two analogous nucleoporins Nup124p and Nup153 wherein the function of a common domain in retrotransposition is conserved.
Figures







Similar articles
-
Multiple conserved domains of the nucleoporin Nup124p and its orthologs Nup1p and Nup153 are critical for nuclear import and activity of the fission yeast Tf1 retrotransposon.Mol Biol Cell. 2007 Sep;18(9):3692-708. doi: 10.1091/mbc.e06-12-1062. Epub 2007 Jul 5. Mol Biol Cell. 2007. PMID: 17615301 Free PMC article.
-
Nup124p is a nuclear pore factor of Schizosaccharomyces pombe that is important for nuclear import and activity of retrotransposon Tf1.Mol Cell Biol. 1999 Aug;19(8):5768-84. doi: 10.1128/MCB.19.8.5768. Mol Cell Biol. 1999. PMID: 10409764 Free PMC article.
-
Nuclear import of the retrotransposon Tf1 is governed by a nuclear localization signal that possesses a unique requirement for the FXFG nuclear pore factor Nup124p.Mol Cell Biol. 2000 Oct;20(20):7798-812. doi: 10.1128/MCB.20.20.7798-7812.2000. Mol Cell Biol. 2000. PMID: 11003674 Free PMC article.
-
Yeast perspectives on HIV-1 VPR.Front Biosci. 2000 Dec 1;5:D905-16. doi: 10.2741/zhao. Front Biosci. 2000. PMID: 11102318 Review.
-
HIV-1 nuclear import: matrix protein is back on center stage, this time together with Vpr.Mol Med. 1998 Mar;4(3):138-43. Mol Med. 1998. PMID: 9562972 Free PMC article. Review. No abstract available.
Cited by
-
The effects of natural selection across molecular pathways in Drosophila melanogaster.BMC Evol Biol. 2015 Sep 21;15:203. doi: 10.1186/s12862-015-0472-4. BMC Evol Biol. 2015. PMID: 26391223 Free PMC article.
-
Insights into cellular factors that regulate HIV-1 replication in human cells.Biochemistry. 2011 Feb 15;50(6):920-31. doi: 10.1021/bi101805f. Epub 2011 Jan 24. Biochemistry. 2011. PMID: 21218853 Free PMC article. Review.
-
Host-HIV-1 Interactome: A Quest for Novel Therapeutic Intervention.Cells. 2019 Sep 27;8(10):1155. doi: 10.3390/cells8101155. Cells. 2019. PMID: 31569640 Free PMC article. Review.
-
The nuclear pore complex: a new dynamic in HIV-1 replication.Nucleus. 2010 Jan-Feb;1(1):18-22. doi: 10.4161/nucl.1.1.10571. Nucleus. 2010. PMID: 21327100 Free PMC article.
-
Versatility at the nuclear pore complex: lessons learned from the nucleoporin Nup153.Chromosoma. 2005 Nov;114(5):319-30. doi: 10.1007/s00412-005-0019-3. Epub 2005 Nov 12. Chromosoma. 2005. PMID: 16133350 Review.
References
-
- Bodoor, K., Shaikh, S., Enarson, P., Chowdhury, S., Salina, D., Raharjo, W. H., and Burke, B. (1999). Function and assembly of nuclear pore complex proteins. Biochem. Cell Biol. 77, 321–329. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

