Pyruvate:quinone oxidoreductase from Corynebacterium glutamicum: purification and biochemical characterization
- PMID: 15659664
- PMCID: PMC545707
- DOI: 10.1128/JB.187.3.862-871.2005
Pyruvate:quinone oxidoreductase from Corynebacterium glutamicum: purification and biochemical characterization
Abstract
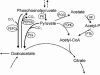
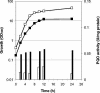
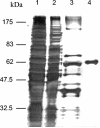
Pyruvate:quinone oxidoreductase catalyzes the oxidative decarboxylation of pyruvate to acetate and CO2 with a quinone as the physiological electron acceptor. So far, this enzyme activity has been found only in Escherichia coli. Using 2,6-dichloroindophenol as an artificial electron acceptor, we detected pyruvate:quinone oxidoreductase activity in cell extracts of the amino acid producer Corynebacterium glutamicum. The activity was highest (0.055 +/- 0.005 U/mg of protein) in cells grown on complex medium and about threefold lower when the cells were grown on medium containing glucose, pyruvate, or acetate as the carbon source. From wild-type C. glutamicum, the pyruvate:quinone oxidoreductase was purified about 180-fold to homogeneity in four steps and subjected to biochemical analysis. The enzyme is a flavoprotein, has a molecular mass of about 232 kDa, and consists of four identical subunits of about 62 kDa. It was activated by Triton X-100, phosphatidylglycerol, and dipalmitoyl-phosphatidylglycerol, and the substrates were pyruvate (kcat=37.8 +/- 3 s(-1); Km=30 +/- 3 mM) and 2-oxobutyrate (kcat=33.2 +/- 3 s(-1); Km=90 +/- 8 mM). Thiamine pyrophosphate (Km=1 microM) and certain divalent metal ions such as Mg2+ (Km=29 microM), Mn2+ (Km=2 microM), and Co2+ (Km=11 microM) served as cofactors. In addition to several dyes (2,6-dichloroindophenol, p-iodonitrotetrazolium violet, and nitroblue tetrazolium), menadione (Km=106 microM) was efficiently reduced by the purified pyruvate:quinone oxidoreductase, indicating that a naphthoquinone may be the physiological electron acceptor of this enzyme in C. glutamicum.
Figures

Similar articles
-
Corynebacterium glutamicum pyruvate:quinone oxidoreductase: an enigmatic metabolic enzyme with unusual structural features.FEBS J. 2024 Oct;291(20):4501-4521. doi: 10.1111/febs.17232. Epub 2024 Jul 30. FEBS J. 2024. PMID: 39080980
-
Pyruvate:quinone oxidoreductase in Corynebacterium glutamicum: molecular analysis of the pqo gene, significance of the enzyme, and phylogenetic aspects.J Bacteriol. 2006 Feb;188(4):1341-50. doi: 10.1128/JB.188.4.1341-1350.2006. J Bacteriol. 2006. PMID: 16452416 Free PMC article.
-
Catalytic properties, molecular composition and sequence alignments of pyruvate: ferredoxin oxidoreductase from the methanogenic archaeon Methanosarcina barkeri (strain Fusaro).Eur J Biochem. 1996 Apr 1;237(1):35-44. doi: 10.1111/j.1432-1033.1996.0035n.x. Eur J Biochem. 1996. PMID: 8620891
-
The pyruvate dehydrogenase complex of Corynebacterium glutamicum: an attractive target for metabolic engineering.J Biotechnol. 2014 Dec 20;192 Pt B:339-45. doi: 10.1016/j.jbiotec.2013.12.019. Epub 2014 Jan 29. J Biotechnol. 2014. PMID: 24486441 Review.
-
Boosting Anaplerotic Reactions by Pyruvate Kinase Gene Deletion and Phosphoenolpyruvate Carboxylase Desensitization for Glutamic Acid and Lysine Production in Corynebacterium glutamicum.Adv Biochem Eng Biotechnol. 2017;159:181-198. doi: 10.1007/10_2016_31. Adv Biochem Eng Biotechnol. 2017. PMID: 27872961 Review.
Cited by
-
Novel, oxygen-insensitive group 5 [NiFe]-hydrogenase in Ralstonia eutropha.Appl Environ Microbiol. 2013 Sep;79(17):5137-45. doi: 10.1128/AEM.01576-13. Epub 2013 Jun 21. Appl Environ Microbiol. 2013. PMID: 23793632 Free PMC article.
-
E1 enzyme of the pyruvate dehydrogenase complex in Corynebacterium glutamicum: molecular analysis of the gene and phylogenetic aspects.J Bacteriol. 2005 Sep;187(17):6005-18. doi: 10.1128/JB.187.17.6005-6018.2005. J Bacteriol. 2005. PMID: 16109942 Free PMC article.
-
Comparative 13C metabolic flux analysis of pyruvate dehydrogenase complex-deficient, L-valine-producing Corynebacterium glutamicum.Appl Environ Microbiol. 2011 Sep;77(18):6644-52. doi: 10.1128/AEM.00575-11. Epub 2011 Jul 22. Appl Environ Microbiol. 2011. PMID: 21784914 Free PMC article.
-
Drought re-routes soil microbial carbon metabolism towards emission of volatile metabolites in an artificial tropical rainforest.Nat Microbiol. 2023 Aug;8(8):1480-1494. doi: 10.1038/s41564-023-01432-9. Epub 2023 Jul 31. Nat Microbiol. 2023. PMID: 37524975 Free PMC article.
-
Flavin redox switching of protein functions.Antioxid Redox Signal. 2011 Mar 15;14(6):1079-91. doi: 10.1089/ars.2010.3417. Epub 2010 Oct 28. Antioxid Redox Signal. 2011. PMID: 21028987 Free PMC article. Review.
References
-
- Abdel-Hamid, A. M., M. M. Attwood, and J. R. Guest. 2001. Pyruvate oxidase contributes to the aerobic growth efficiency of Escherichia coli. Microbiology 147:1483-1498. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Andersson, L., H. Borg, and M. Mikaelsson. 1972. Molecular weight estimations of proteins by electrophoresis in polyacrylamide gels of graded porosity. FEBS Lett. 20:199-202. - PubMed
-
- Birnboim, H. C. 1983. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 100:243-255. - PubMed
-
- Blake, R., and L. P. Hager. 1978. Activation of pyruvate oxidase by monomeric and micellar amphiphiles. J. Biol. Chem. 253:1963-1971. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

